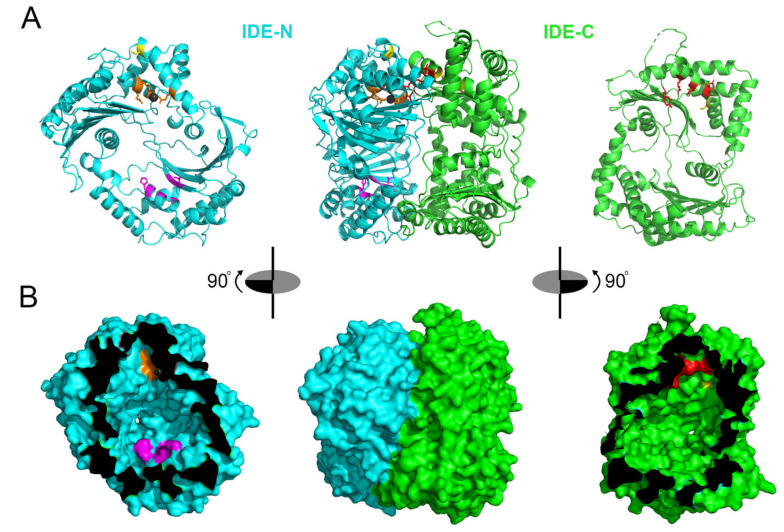
Figure 2.
Structure of human IDE and regions targeted by different inhibitors. (A,B), Illustration of a single monomer of IDE (center) and IDE-N (left, cyan) and IDE-C (right, green) depicted in ribbon (A) and surface (B) representations. Zinc-binding and catalytic residues within the active site of IDE-N are depicted in orange, with the zinc atom shown as a gray sphere. Residues within IDE-C that make up the second portion of IDE’s bipartite active site are shown in red. Cysteine residues targeted by thiol-modifying inhibitors are shown in yellow. The distal exosite is shown in magenta. In (B), note that the portions of IDE-N and IDE-C that are adjacent when the protease is in the closed conformation are depicted in black. Figures generated in Pymol [169] from PDB 2G54 [20].