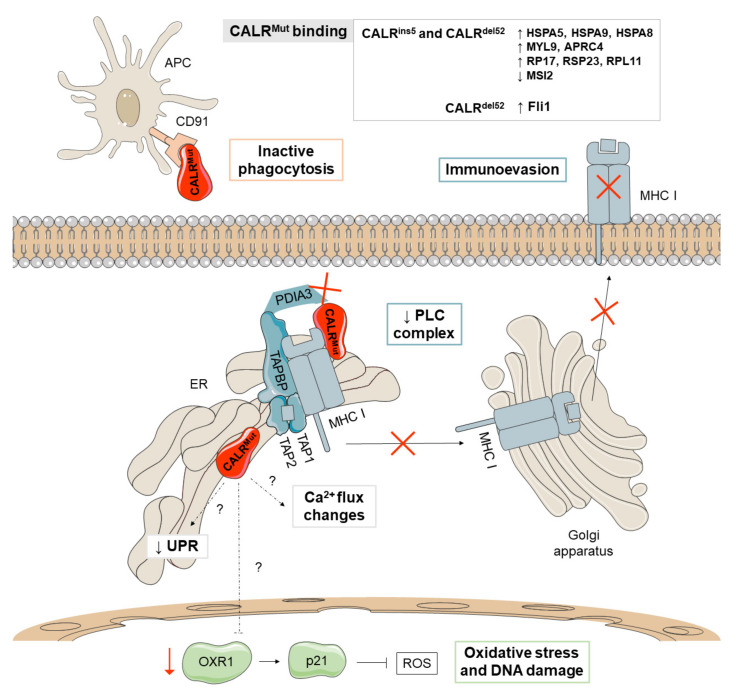
Figure 3.
Major non-canonical mechanisms derived from CALRMut. CALRMut (depicted in red) shows different binding affinities for proteins implicated in the unfolding protein response (UPR) (HSPA5, HSPA9, and HSPA8), proteins of the cytoskeleton (MYL9 and APRC4), and ribosomal proteins (RP17, RSP23, and RPL11), as well as reduced binding to MSI2, a transcriptional regulator that target genes mainly involved in cell cycle regulation. Additionally, CALRMut seems to reduce the activation of the pro-apoptotic pathway of the UPR and increases oxidative stress and DNA damage through the downmodulation of oxidation resistance 1 (OXR1). CALRMut also shows decreased binding affinities for PDIA3 and has a loss-of-function effect on the peptide loading complex (PLC), which mediates the loading of cellular antigens onto major histocompatibility complex class I (MHC-I) molecules, favoring immunoevasion. Mutations in CALR increase the secretion of the protein and bind to CALR receptors in antigen presenting cells (APCs), limiting their ability to phagocytize wild-type CALR-exposing cancer cells. The main differences between the phenotypes observed in patients with type 1 (del52) and type 2 (ins5) mutations have been attributed to thrombopoietin receptor (TPOR)-independent cytosolic calcium fluxes and the binding affinity for the transcription factor FLI1.