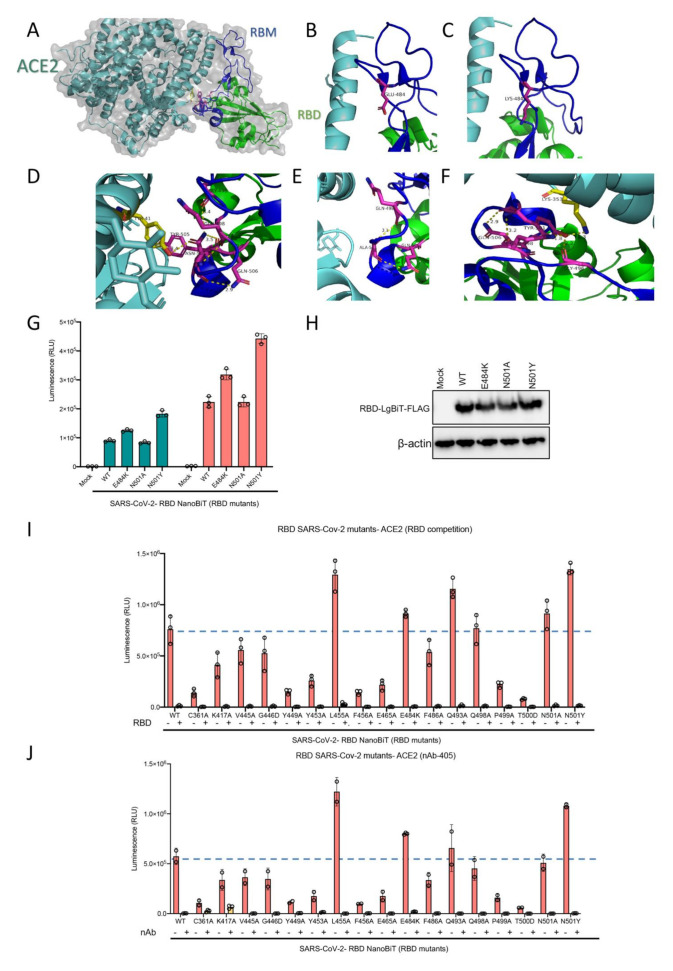
Figure 6.
(A–F) Visualization of RBD N501 and E484 mutations. (A) Overall structure of RBD and ACE2 interaction. (B) The loop structure in RBD containing E484 residue. (C) Predicted structure following E484 residue mutation to lysine which would not significantly change the nature of its interaction in the loop structure of RBD, (D) The N501 residue and its interacting residues in RBM and ACE2. (E) Predicted structure when the N501 residue mutates to alanine which would eliminate ACE2 interaction but preserve internal RBM interactions. (F) Predicted structure when the N501 residue mutates to tyrosine which would preserve and slightly boost internal interactions, and introduce a stronger interaction site with ACE2 compared to the wild type residue. (G) ACE2 demonstrating altered binding affinity of various mutants of RBD-LgBiT-mutants. (H) Immunoblot of RBD-LgBiT mutant expression from the cell lysates of transfected HEK293T cells. β-actin is shown as control. Competition biosensor assay with LgBiT-RBD mutants which maintained any binding capacity pre-incubated with (I) recombinant RBD protein or (J) neutralizing antibody to examine therapeutic implications of various mutants.