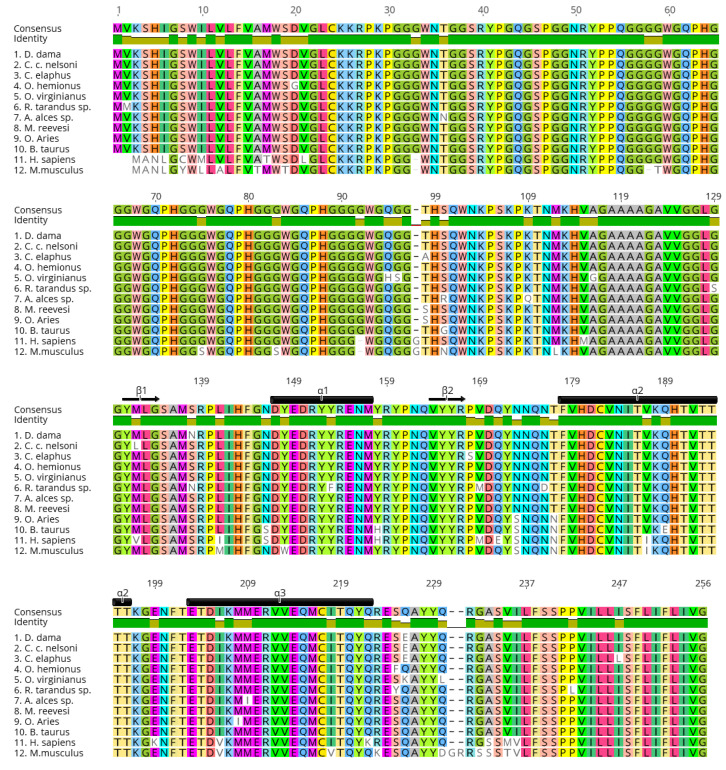
Figure 3.
Cervid prion protein sequence alignment showing conserved homology between species. Protein alignment was performed in Geneious v10.2.6 (https://www.geneious.com) (accessed on 15 December 2020) using the ClustalW algorithm. Amino acid numbering is based on the consensus sequence. Amino acid variants were added manually to each sequence and are shown in white boxes. NCBI accession numbers used in this alignment: (1) QAU19527.1, (2) ABW79881.1, (3) QAU19537.1, (4) AAO91945.1, (5) QKI87491.1, (6) AAT77253.1, (7) QHZ32187.1, (8) AGU92564.1, (9) ABA08026.1, (10) BAI50003.1, (11) BCK59655.1, (12) CAJ18553.1. β1, β2: first, second beta-strand; α1, α2, α3: first, second and third alpha-helix (based on mouse PrP numbering [136]). Refer to Table 1 for cervid species names in sequences 1–8. Non-cervid species names for sequences 9–12: O. = Ovis, B. = Bos, H. = Homo and M. = Mus.