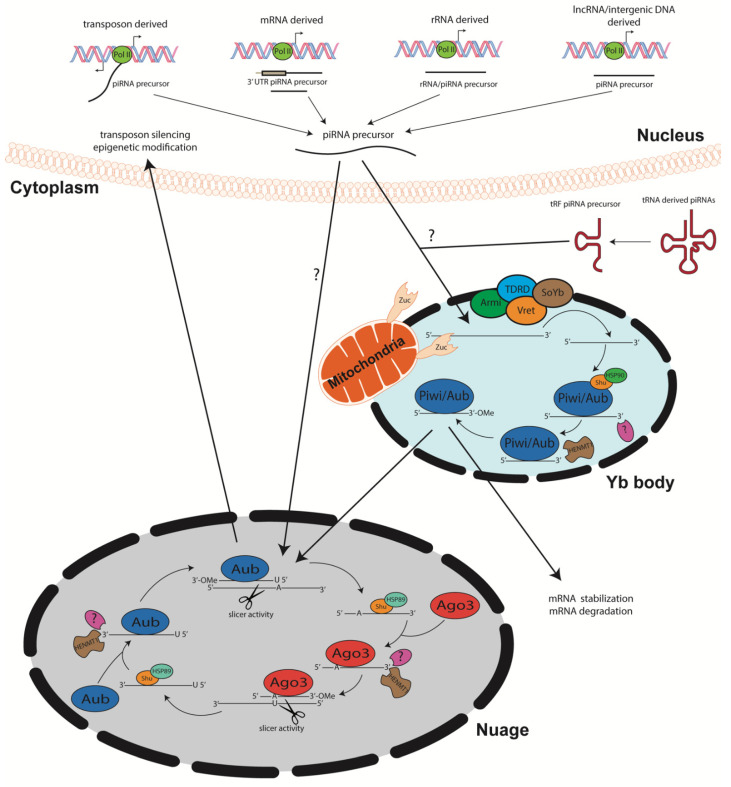
Figure 1.
piRNA biogenesis Precursor piRNAs within the nuclear compartment are generated from sense and antisense transposon sequences, 3′UTR of messenger RNAs (mRNAs), ribosomal RNAs (rRNAs), long noncoding RNAs (lncRNAs) and intergenic DNA sequences. Furthermore, piRNAs can be derived from transfer RNA (tRNA), specifically transfer RNA related fragments (tRFs) that are processed in the cytoplasm. During primary biogenesis within the Yb body (shown to occur in somatic cells) precursor piRNA is associated with various processing proteins including Tutor domain containing protein (TDRD), Armitage (Armi), Vreteno (Vret), and Sister of Yb (SoYb), though their specific functions during piRNA biogenesis is not well understood. piRNA precursors are processed by mitochondria membrane bound protein Zucchini (Zuc) into small segments. Shutdown (Shu) and Heat shock protein 90 (HSP90) shuttle piRNAs onto Piwi proteins. An unknown endonuclease further processes the 3′ end of the piRNA before piRNA methyltransferase Hen1 (HENMT1) adds a 3′-OMe group. Mature piRNAs enter the ping pong cycle to produce progeny piRNA, re-enter the nucleus for transposon silencing and epigenetic modification, or enter the cytoplasm to play roles in mRNA transcript stabilization or degradation. The ping-pong cycle occurs in the Nuage (shown to occur in germ cells), where antisense piRNAs bound to Aubergine (Aub) can bind to sense piRNA precursor molecules, allowing the RNAse H-like slicer activity to process the precursor piRNA into a smaller segment. Shu and HSP89 shuttle processed piRNAs onto Argonaute 3 (Ago3). An unknown protein processes the 3′ end, and subsequently HENMT1 adds the characteristic 3′-OMe that is required for piRNA stabilization. Mature, sense, Ago3 bound piRNAs can continue to process more antisense precursor piRNAs that will bind to Aub and continue the cycle of piRNA secondary biogenesis. The mature, stable piRNAs synthesized from the ping-pong cycle can then enact various piRNAs function in the nucleus and/or in the cytoplasm.