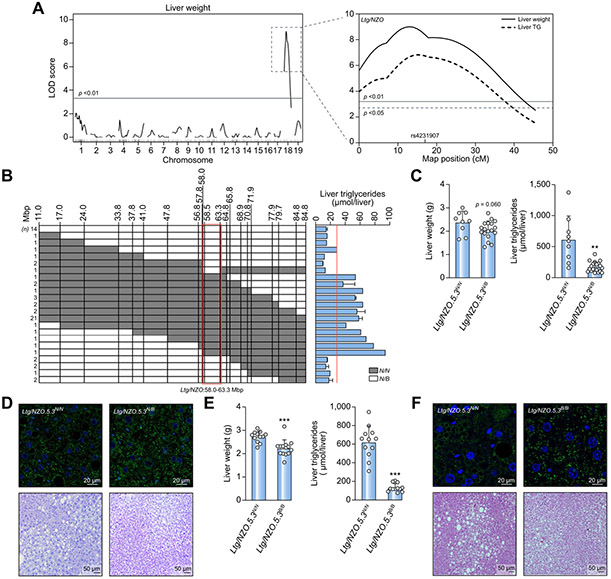
Fig. 1. Identification of a susceptibility locus for fatty liver on chromosome 18.
(A) Genome-wide linkage analysis of (NZOxB6)N2 population for liver weight revealed a QTL on chromosome 18. LOD score curves for liver weight and triglycerides. Solid horizontal (p <0.01) and dashed lines (p <0.05) indicate threshold of significance calculated with 1,000 permutations. (B) Identification of the critical region of Ltg/NZO (red box) and liver triglycerides in (NZOxB6)N2 mice. (C,E) Liver weight and triglyceride concentrations of RCS Ltg/NZO.5.3N/N (n = 9) and Ltg/NZO.5.3N/B (n = 18) (F4.N10) as well as Ltg/NZO.5.3N/N (n = 12) and Ltg/NZO.5.3B/B (n =12) (F8.N9). Data analyzed by unpaired t test with Welch's correction and presented as mean ± SD. **p <0.01; ***p <0.001. (D,F) Histology of liver sections from 12-week-old mice stained for PLIN2 (green) and TO-PRO (blue, nuclei) as well as H&E staining. B6, C57BL/6; Ltg/NZO, liver triglycerides from NZO alleles; NZO, New Zealand obese; QTL, quantitative trait loci.