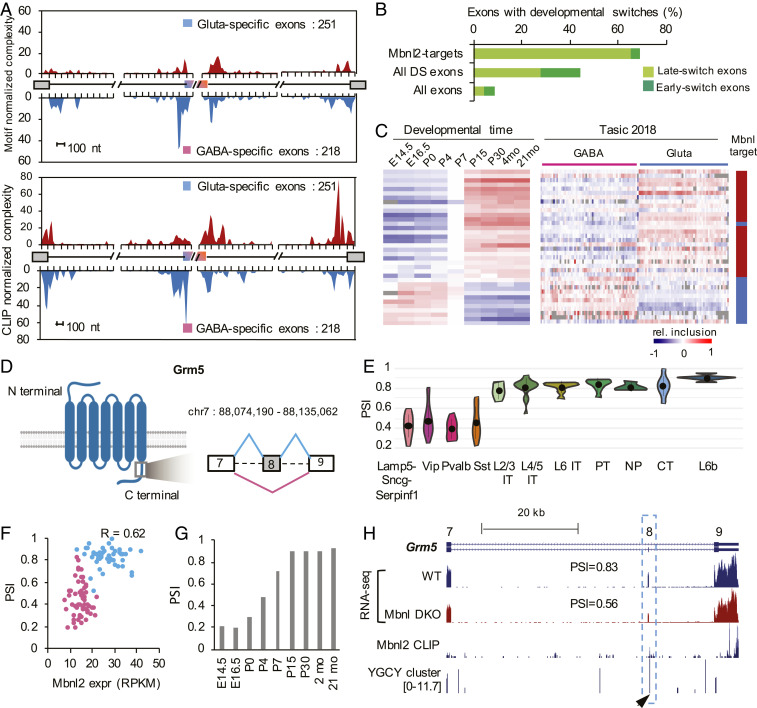
Fig. 7.
Mbnl2 drives a glutamatergic neuron-specific splicing profile. (A) RNA maps showing enrichment of Mbnl binding sites (predicted YGCY clusters at the top and Mbnl2 CLIP tags at the bottom) in the upstream intron of cassette exons with GABAergic neuron-specific inclusion and in downstream intron of cassette exons with glutamatergic neuron-specific inclusion. (B) DS exons regulated by Mbnl are enriched in late-switch exons during cortical development as compared to all DS exons or all cassette exons. Exons showing early or late developmental splicing switches were obtained from Weyn-Vanhentenryck et al. (12). (C) Comparison of splicing profiles of DS exons regulated by Mbnl during cortical development (12) (Left) and across 117 neuronal cell types from Tasic 2018 (Right). Exons activated or repressed by Mbnl are indicated in red and blue, respectively, in the scale bar on the right. (D–H) Regulation of Grm5 exon 8 by Mbnl2 is correlated with glutamatergic neuron-specific inclusion. (D) Schematic diagram of Grm5 protein domain architecture and the region encoded by exon 8 with preferential inclusion in glutamatergic neurons. (E) Violin plots showing Grm5 exon 8 inclusion in glutamatergic and GABAergic neuronal subclasses. Median and the interquantile range are indicated. (F) Correlation of exon inclusion and Mbnl2 expression across 117 neuronal cell types. Glutamatergic and GABAergic neuronal cell types are colored in blue and pink, respectively. Pearson’s correlation coefficient is indicated. (G) Grm5 exon 8 inclusion during cortical development. (H) Mbnl2 bind to the downstream intron of Grm5 exon 8, as shown by Mbnl2 CLIP data and predicted Mbnl-binding YGCY clusters (arrowhead). Exon inclusion in the cortex is reduced upon depletion of Mbnl1/2 (Mbnl DKO).