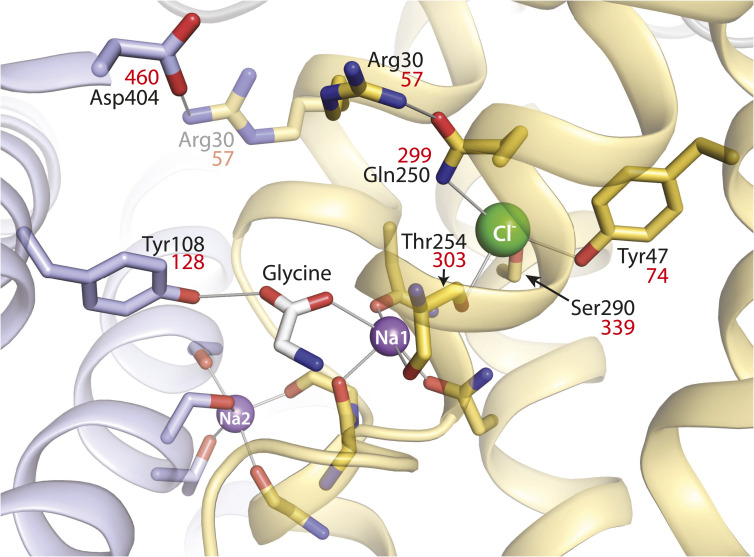
Fig. 1.
LeuT-E290S with bound Cl−, showing interaction networks proposed to account for the effect of ligand binding on conformational change. Outward-occluded conformation of LeuT E290S, a Cl−-dependent mutant [PDB 4HOD (36)]. Bundle helices are in gold and scaffold helices in light blue, and key residues are shown as sticks. A leucine molecule in the substrate site was mutated to glycine and is shown as sticks with the C atoms in white. Ions are shown as spheres for sodium (purple) and chloride (green). Two ligand-dependent networks of interaction connect the scaffold and bundle domains across the extracellular pathway, as indicated by thin gray lines. One of these pathways involves the carboxyl group of substrates connecting Tyr108 from the scaffold domain and Na1 in the bundle domain (28). The other network involves an ion pair between Asp404 (scaffold) and Arg30 (bundle). Arg30 is shown with two rotamers, one (from the LeuT E290S structure) interacting with Gln250 in the Cl− binding site and the other (semitransparent) interacting with Asp404. For clarity, only residues participating in the substrate- and Cl−-dependent conformational changes and the Cl− binding site are numbered. Na+ coordinating residues (not labeled) include Ala22, Asn27, Thr254, and Asn286 (Na1) and Gly20, Val23, Ala351, Thr354, and Ser355 (Na2) (19). Residue numbers in red are the corresponding positions in GlyT1b (43).