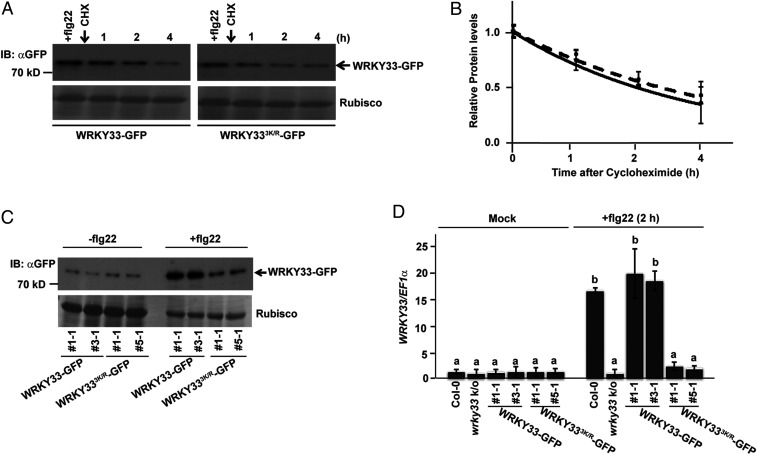
Fig. 2.
SUMOylation is essential for transcriptional activation of WRKY33. (A) SUMOylation does not affect WRKY33 protein turnover rates. Twelve-day-old seedlings of both WRKY33 and WRKY333K/R transgenic lines were first treated with flg22 for 2 h for induction of the respective proteins and then cycloheximide (200 μM) treatment was done for the indicated timepoints. The samples obtained were immunoblotted with αGFP for detecting WRKY33-GFP. Ponceau stained Rubisco was used as the loading control. (B) Quantification of the bands shown above were plotted. Relative protein abundances were normalized to the loading control (Rubisco) and to their respective levels at time 0 h. Results from two independent replicates were averaged and shown with error bars denoting SD. An exponential best-fit curve was fitted through the data points. (C) WRKY33 protein induction is attenuated in WRKY333K/R transgenics. Total protein from 12-d-old seedlings of WRKY33 and WRKY333K/R transgenics were treated with −/+flg22 (1 µM) and then samples were immunoprecipitated with anti-GFP (IP: αGFP) beads. The samples were immunoblotted with anti-GFP (IB: αGFP) antibodies. Rubisco was used as the loading control. (D) WRKY33 SUMOylation is essential for its transcriptional activation upon flg22 treatment. Twelve-day-old seedlings of the different genotypes indicated were treated with 1 μM flg22 for 2 h and WRKY33 expression was compared. The mock-treated seedling for each genotype was taken as controls. Bars represent means ± SD from two independent biological replicates. Col-0 was used as the reference and normalization was done using EF1α. Genotypes with different letters were significantly different from others (two-tailed Student’s t test; P < 0.05).