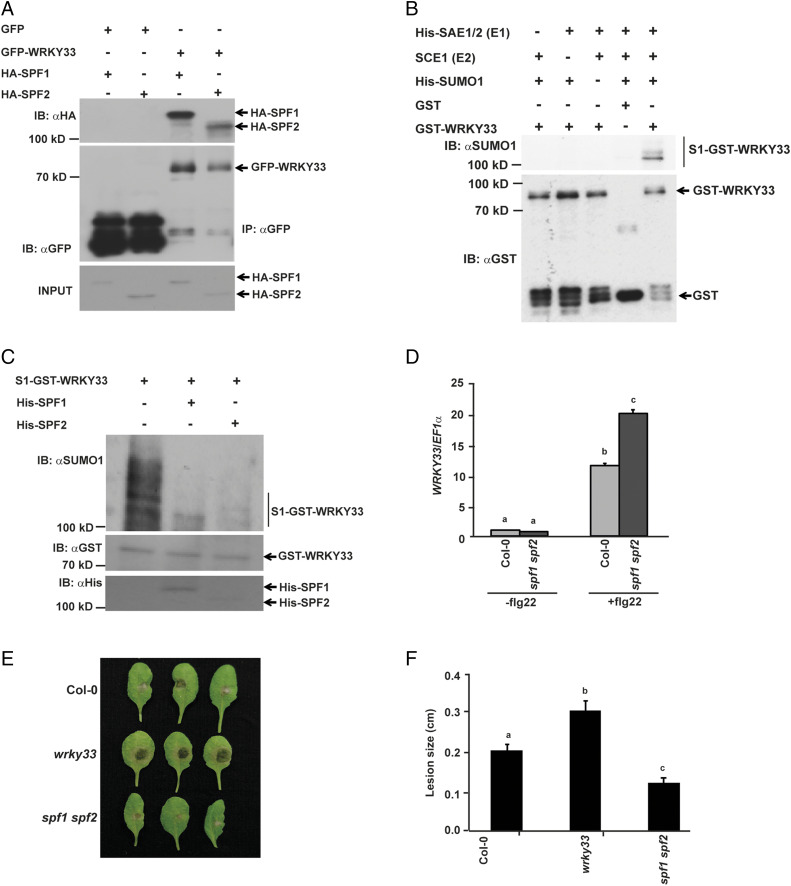
Fig. 3.
WRKY33 deSUMOylation is regulated by SPF1 and SPF2 SUMO proteases. (A) WRKY33 interacts with SPF1 and SPF2. Total protein from N. benthamiana leaves transiently expressing GFP-WRKY33 either with HA-SPF1 or HA-SPF2 was subjected to IP with anti-GFP beads (IP: αGFP) and immunoblotted with anti-HA (IB: αHA) for SCE1-HA and anti-GFP (IB: αGFP) for GFP/GFP-fusion proteins. Total protein extracts were probed with anti-HA antibody (HA-SPF1/HA-SPF2 input) for equal HA-fusion proteins. (B) WRKY33 can be SUMOylated under in vitro conditions. Recombinantly expressed GST-WRKY33 was incubated with His-SAE1, SCE1, and His-SUMO1 at 30 °C for 2 h. Besides the GST-only control, independent reactions were set up as negative controls with one component missing to ensure that all components are essential for WRKY33 SUMOylation. Reactions were stopped with 4× SDS buffer and immunoblotted with anti-GST (IB: αGST) and anti-SUMO1/2 (IB: αSUMO1) antibodies. (C) About 10 μg of SUMOylated GST-WRKY33 was incubated either with His-SPF1 or His-SPF2 for 16 h and the reactions were stopped by adding 4× dye. The samples were immunoblotted with αGST for GST-WRKY33, αHis for His-SPF1 and SPF2, and with αSUMO1 for detecting SUMOylated WRKY33. (D) WRKY33 expression is higher in spf1 spf2 mutant compared to Col-0. WRKY33 expression was analyzed in 12-d-old seedlings of the different genotypes indicated before and after treatment with 1 µM flg22. Data presented are means ± SD from two independent biological replicates and normalized to EF1α expression. A two-tailed Student’s t test showed that WRKY33 expression was significantly different in genotypes with different letters (P < 0.05). (E) spf1 spf2 mutant is more resistant to B. cinerea infection. Fully expanded leaves from 4-wk-old plants of Col-0, wrky33 k/o and spf1 spf2 were excised and drop-inoculated with 5 µL B. cinerea spore suspension. Photographs were taken 72 h postinfection and quantified in F. (F) The data represent the means ± SE of lesion sizes from 20 to 25 individual leaves per genotype. A two-tailed Student’s t test was performed among means of lesion sizes. Genotypes with different letters were significantly different from others (P < 0.05).