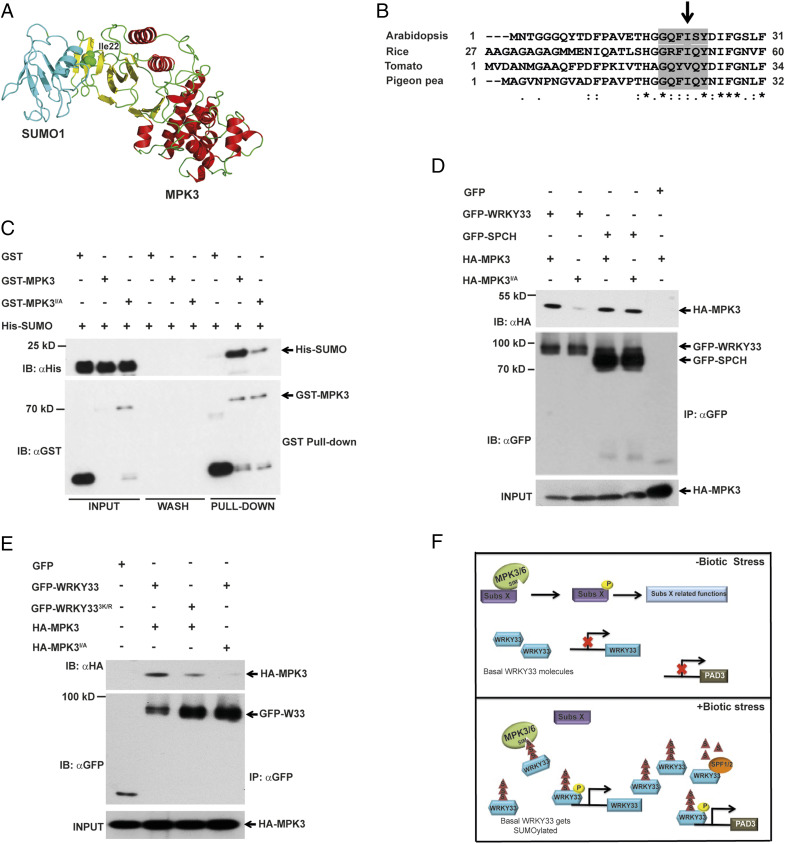
Fig. 6.
SUMO–SIM module facilitates WRKY33-MAPK Interaction. (A) A structural snapshot of MPK3–SUMO1 protein complex model highlighting the docking of SUMO1 onto MPK3 N-terminal region containing the SIM site (Ile22). MPK6 structure (PDB ID: 5CI6) was used as a template for MPK3 model (red and yellow). SUMO1 (PDB ID: 4WJO) is in turquoise. MPK3 Ile22 as green spheres. (B) Arabidopsis MPK3 Ile22 is conserved among its homologs from rice, tomato and pigeon pea. SIM is highlighted in gray. (C) MPK3 SIM site is required for interaction with SUMO. An in vitro pull-down assay exhibited that MPK3 SIM mutant (MPK3I22A) interacted weakly with SUMO. Purified GST-MPK3 and GST-MPK3I22A were incubated with His-SUMO for 1 h and eluted after unbound proteins were washed. Input, wash, and elution (pull-down) were immunoblotted with anti-GST (IB: αGST) and anti-His (IB: αHis) antibodies. GST was used as a negative control. (D) MPK3 SIM site is required for WRKY33 but not SPCH. Total protein from N. benthamiana leaves transiently expressing GFP-WRKY33/GFP-SPCH with HA-MPK3/HA-MPK3I22A was immunoprecipitated with anti-GFP beads (IP: αGFP) and immunoblotted with anti-HA (IB: αHA) for HA-MPK3/HA-MPK3I22A and anti-GFP (IB: αGFP) for GFP/GFP-fusion proteins. Extracts were probed with anti-HA antibody (HA-MPK3 input) for equal MPK3/MPK3I22A. (E) WRKY33-MPK3 interaction is reliant on SUMO–SIM module. Total protein from N. benthamiana leaves transiently expressing GFP-WRKY33/GFP-WRKY333K/R with either HA-MPK3/HA-MPK3I22A was subjected to IP with anti-GFP beads (IP: αGFP) and immunoblotted with anti-HA (IB: αHA) for HA-fusion proteins and anti-GFP (IB: αGFP) for GFP/GFP-fusion proteins. Extracts were probed with anti-HA antibody (HA-MPK3/HA-MPK3I22A input) for equal HA-fusion proteins. (F) A simplistic model showing that WRKY33 SUMOylation is an early plant response upon pathogen infection and allows WRKY33 to be quickly and specifically identified by MAPKs SIM site via the SUMO–SIM module. In absence of stress, basal-level WRKY33 is unperturbed and MAPKs interact and phosphorylate substrate X (Subs X), resulting in regulation of its functions. However, during biotic stress, basal-level WRKY33 is rapidly SUMOylated to be selectively and quickly identified by MPK3/6 via the SUMO–SIM module for phosphorylation and activation, resulting in activation of downstream genes (PAD3). SPF1/2 regulates WRKY33 SUMO levels. Yellow circles with “P” are phosphorylation; triangles with “S” are SUMOylation.