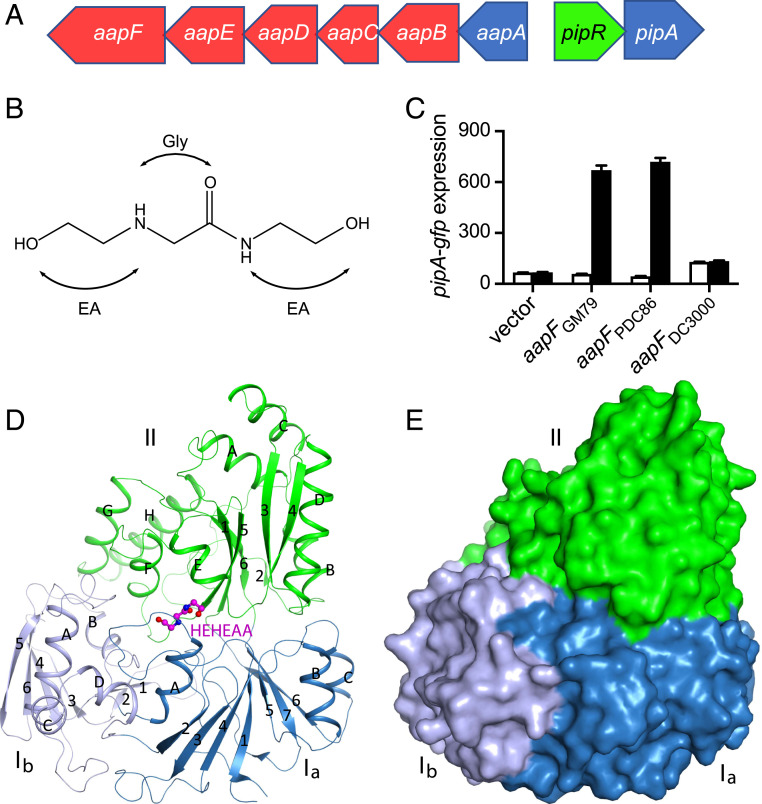
Fig. 1.
The Pseudomonas GM79 pipR genomic region, HEHEAA chemical structure, AapF activity analysis, and structure of SBPPDC86-HEHEAA complex. (A) The pipR (green) genomic region. Genes coding for the predicted ABC-type transporter are in red, and genes coding for peptidases are in blue. (B) Chemical structure of HEHEAA with the ethanolamine (EA) and glycine (Gly) substructures indicated. (C) SBPPDC86, but not SBPDC3000, can substitute for SBPGM79. Data are pipA-gfp fluorescence in a GM79 aapF mutant harboring plasmids with the indicated aapF gene and a pipA-gfp reporter. Cells were grown with (black bars) or without (white bars) 100 nM HEHEAA. The pipA-gfp expression is in relative fluorescence units (RFU) per optical density (OD600) of four biological replicates, and the error bars represent the SDs of the mean. (D) Ribbon diagram of the overall structure of SBPPDC86-HEHEAA complex. The three domains Ia, Ib, and II are labeled and colored in sky blue, gray blue, and green, respectively. β-strands and α-helices are labeled with numbers and letters, respectively. The ligand HEHEAA is shown as ball-and-stick model in magenta. (E) Surface representation of SBPPDC86 with the same color scheme as in D. The HEHEAA ligand is completely buried inside the structure. The structure figures were produced with PyMOL (www.pymol.org).