Fig. 4.
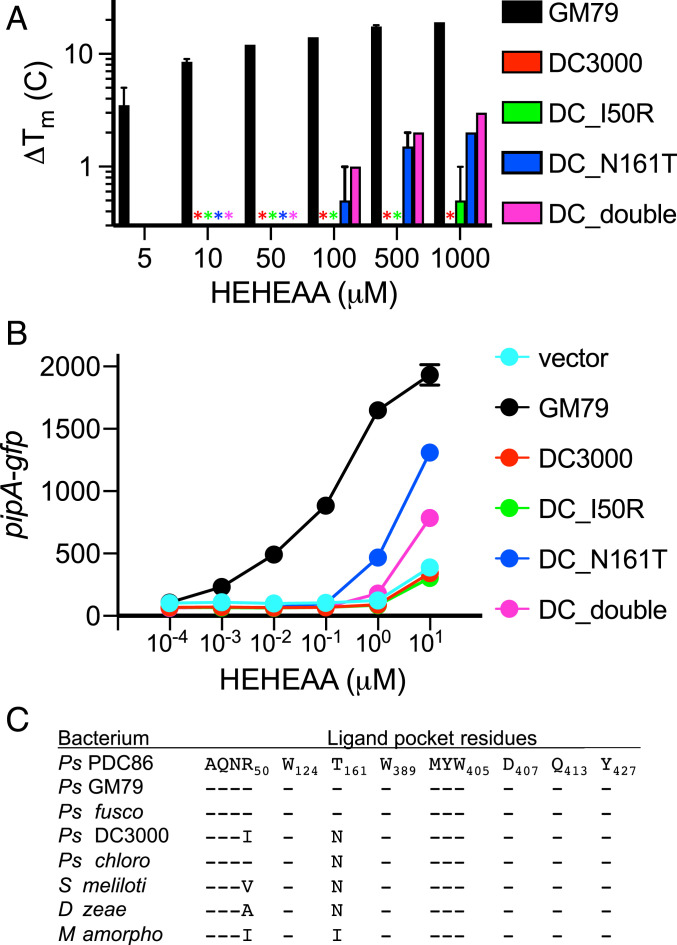
Not all AapF homologs bind HEHEAA. (A) Variant, but not wild-type, AapFDC3000 polypeptides bind HEHEAA in thermal shift experiments. The mean melting temperature changes (ΔTm) of three technical replicates for each of two biological replicates for each indicated AapF are shown and the error bars represent the range of values. The stars indicate there was no thermal shift. Thermal shift raw data are plotted in SI Appendix, Fig. S7. (B) Genes encoding a specific SBPDC3000 variant (N161T) can complement a GM79 aapF deletion mutant. Data are GFP fluorescence in a GM79 aapF mutant harboring the indicated plasmids. Plasmids used are pJN-PpipA-gfp (vector), pJN-aapFGM79PpipA-gfp (GM79), pJN-aapFDC3000PpipA-gfp (DC3000), pJN-aapFDC_I50RPpipA-gfp (DC_I50R), pJN-aapFDC_N161TPpipA-gfp (DC_N161T), and pJN-aapFDC_doublePpipA-gfp (DC_double). Data are mean RFU per optical density (OD600) of four biological replicates, and the error bars represent the SD of the mean. (C) The ligand-binding pocket residues in AapF homologs from eight bacterial species. The AapF homologs used for comparisons are provided in SI Appendix, Table S5. Residue comparisons are relative to the SBPPDC86 (Top), and a dash (-) indicates that the corresponding residue is the same as in SBPPDC86.