Figure 2. De novo sequence determination of nucleobase peptides with up to four nucleobases per compound is possible using tandem MS/MS and sequencing software.
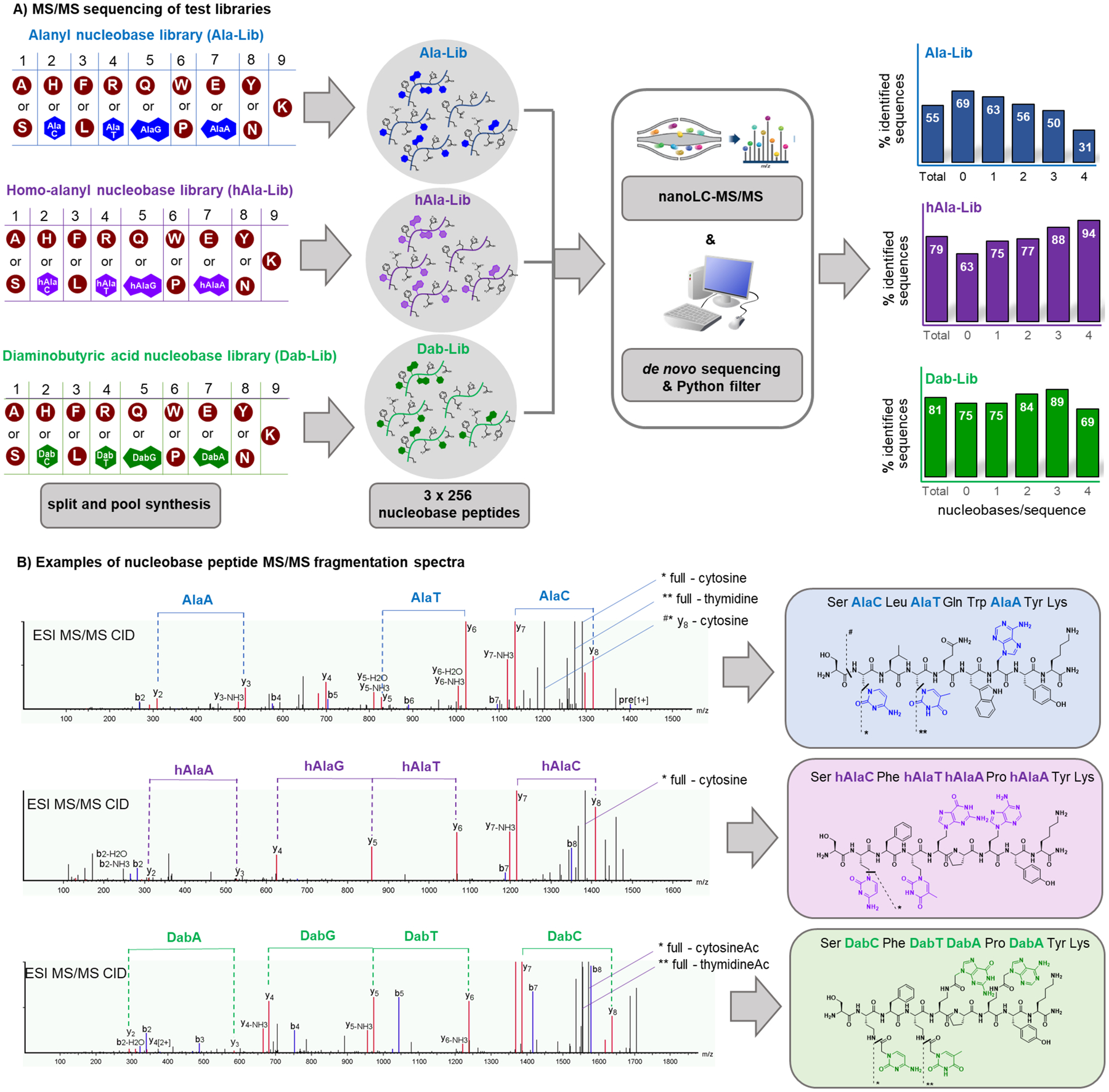
A) Three combinatorial libraries with the three nucleobase monomer sets (alanyl = blue; homoalanyl = purple; Dab = green) and canonical amino acid residues were synthesized and analyzed by nanoLC-MS/MS. De novo sequencing was performed with the software PEAKS 8.5 and the final list was filtered for sequences fitting the correct library design. The percentage of identified total sequences and sequences containing different numbers of nucleobases is reported on the graphs on the right. Expected total sequences with N nucleobases: 16 for N = 0, 64 for N = 1, 96 for N = 2, 64 for N = 3, 16 for N = 4. B) Exemplary MS/MS spectra of nucleobase peptides from the three libraries. Fragmentations of nucleobase side chains are shown.
