Figure 5. A nucleobase peptide with nanomolar affinity for RNA-hairpins was identified by affinity selection.
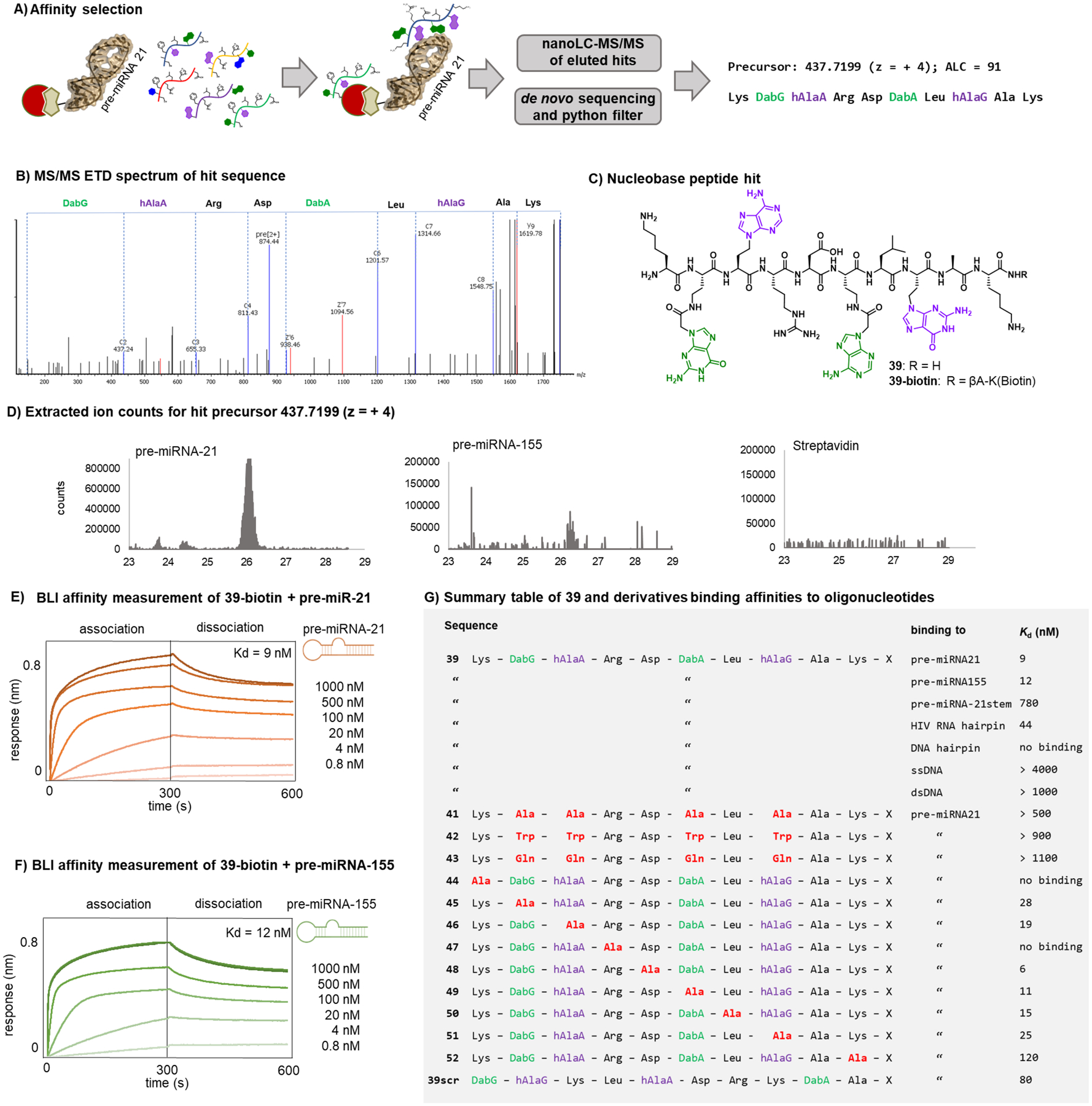
A) Schematic representation of the magnetic bead-based affinity selection with nucleobase peptide library (streptavidin coated magnetic beads = red; biotinylated pre-miRNA = brown). Total immobilized RNA: 100 pmol; [library] = 1 mM; [individual library member] = 10 pM; incubation buffer: Tris 20 mM, pH = 7.5, NaCl 300 mM, MgCl2 20 mM, Tween 0.05 %, Yeast RNA 0.2 mg/mL, Biotin 2 mM; B) ETD spectrum of hit sequence identified by affinity selection. C) structure of hit sequence. D) extracted ion counts for the precursor mass (437.72; z = + 4) in the MS chromatograms of the affinity selection with pre-miRNA-21 (Biotin- UGUCGGGUAGCUUAUCAGACUGAUGUUGACUGUUGAAUCUCAUGGCAACACCAGUCGAUGGGCUGUCUGACA), pre-miRNA-155 (Biotin-CUGUUAAUGCUAAUCGUGAUAGGGGUUUUUGCCUCCAACUGACUCCUACAUAUUAGCAUUAACAG) and the negative control streptavidin beads only. E) BLI association/dissociation of pre-miRNA-21 to immobilized 39-biotin. F) BLI association/dissociation of pre-miRNA-155 to immobilized 39-biotin. G) Summary table of 39 and derivatives binding to RNA and DNA oligonucleotides. The table lists also the affinities towards pre-miRNA-21 of alanine scan mutants and nucleobase mutants of 39. X = -βAla-Lys(Biotin)-CONH2
