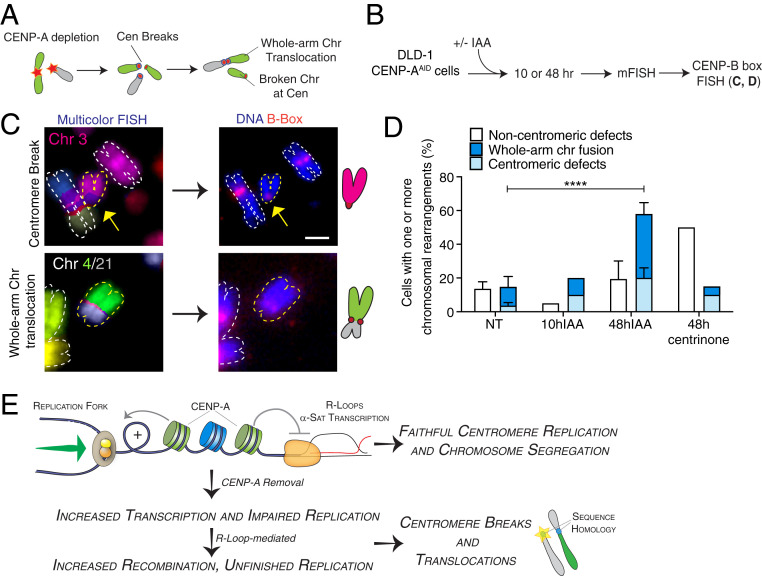
Fig. 5.
Centromere instability caused by CENP-A depletion during DNA replication initiates chromosome rearrangements and translocations in the ensuing cell cycles. (A) Illustration of the formation of centromeric chromosomal rearrangements after CENP-A depletion due to breaks generated during the previous cell cycle and repaired by nonallelic homologous recombination. (B) Schematic representation of the multicolor-FISH experiments shown in C and D. (C) Representative images of mFISH and centromeric FISH (CENP-B box probe) of DLD-1 chromosome spreads showing chromosomal aberrations occurring at centromeres after 48 h IAA. Schematics of the observed events are also shown. (Scale bar, 2 µm.) (D) Quantification of the percentage of metaphase spreads showing chromosomal rearrangements by mFISH after CENP-A depletion or centrosome depletion in DLD-1 cells. n = 3 (NT and 48 h IAA) or 1 (10 h IAA and 48 h 0.2 μM centrinone). The bars represent the SEM. χ2 test: ****P < 0.0001. (E) Model for maintenance of centromeric DNA stability mediated by CENP-A–containing chromatin. See text for details.