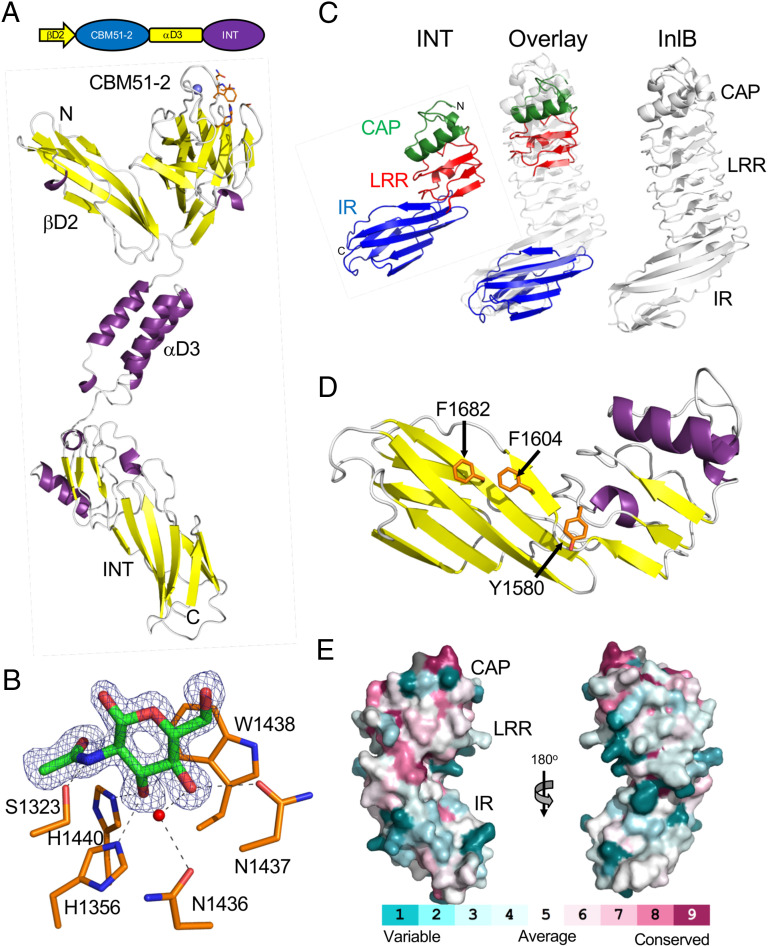
Fig. 4.
Structure of the C terminus of ZmpB. (A) Modular schematic of the crystallized and modeled βD2/INT protein according to Fig. 1 shown with a cartoon representation of the unliganded βD2/INT crystal structure. The N and C termini are indicated, and a bound calcium ion is shown as a blue sphere. The carbohydrate binding region of the protein is indicated by the orange amino acid side chains that are shown as sticks. (B) A close-up of the CBM51-2 carbohydrate binding site in the βD2/INT GlcNAc complex. The electron density of the bound GlcNAc residue (green sticks) is shown as a blue mesh σa-weighted Fo-Fc omit map contoured at 3σ. Residues involved in the interaction are shown as an orange stick, hydrogen bonds as dashed lines, and a coordinated water as a red sphere. (C) Comparison of the INT domain, and its subdomains, with Internalin B (InlB) from Listeria monocytogenes (PDB ID 1H6T). The cap (CAP) subdomain is shown in green, the leucine-rich repeat (LRR) in red, and the Ig-like fold (IR) in blue. The overlay was generated by separately overlapping the CAP/LRR and IR fragments with InlB. (D) Conservation of the INT surface residues with ∼163 nonredundant INT homologs with greater than 30% amino acid sequence identity and 75% sequence coverage. The conservation was mapped onto the INT structure using CONSURF (72). (E) Structure of the INT domain showing distinctive solvent-exposed aromatic amino acid residues.