Fig. 1.
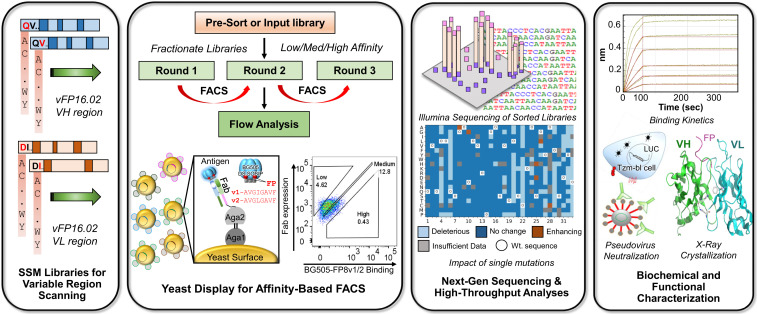
Experimental workflow for comprehensive functional analysis of all possible single mutations in an HIV-1 bNAb. SSM libraries were designed for VH and VL regions of the anti–HIV-1 FP bNAb vFP16.02. SSM libraries were cloned into yeast Fab surface display libraries and screened by FACS to determine the functional impact of each possible single mutation. Single-mutation display libraries were sorted for their affinity against a BG505 DS-SOSIP HIV-1 Env trimer with two different FP sequences. Sorted yeast libraries were prepped for NGS to enable quantitative variant tracking across three screening rounds; bioinformatic analyses of NGS data were used to interpret the functional impact of each possible amino acid mutation. Selected variants were characterized for neutralization activity and affinity against soluble FP and against HIV-1 Env. Structural analyses were performed to understand the mechanistic basis of anti–HIV-1 FP bNAb improvement.