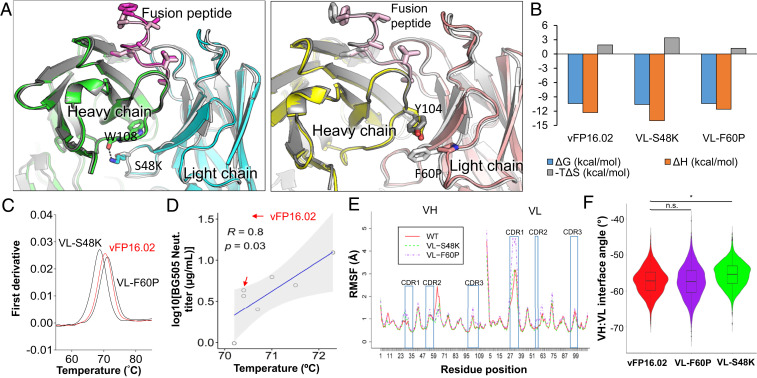
Fig. 6.
Structural basis of enhanced HIV-1 fusion peptide recognition. (A) Ribbon representation of an X-ray crystal structure of VL-S48K (Left) and VL-F60P (Right) in complex with FP8v1 and structurally superposed on wild-type vFP16.02 (shown in gray). (B) Thermodynamic binding interaction parameters obtained by isothermal titration calorimetry for VL-S48K, VL-F60P, and vFP16.02 in complex with the BG505-FP8v1.DS.SOSIP trimer. (C) nanoDSF measurements of melting temperature (Tm) for single vFP16.02 variants. VL-S48K and VL-F60P had opposing effects on the Tm of vFP16.02 (shown in red for comparison). (D) Correlation between Tm and BG505 neutralization titers for single-mutation neutralizing variants that were predicted to bind to BG505-FP8v1.DS.SOSIP. Shaded area indicates 95% confidence limits. (E) Average RMSF plot for three independent molecular dynamics simulation performed for vFP16.02, VL-S48K, and VL-F60P. (F) VH:VL interface angle distribution throughout molecular dynamics simulations, calculated using ABangle. VL-S48K showed a statistically significant change in interface angle with respect to vFP16.02 and VL-F60P with *P < 0.0001 using Mann–Whitney U test. vFP16.02 and VL-F60P interface angles were not statistically significant. n.s., not significant.