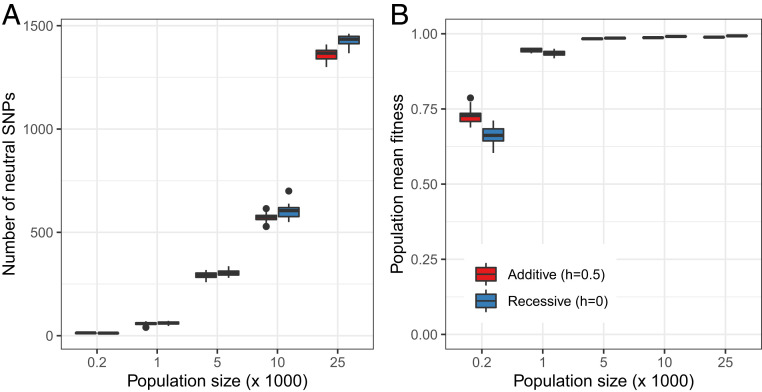
Fig. 3.
Forward in time simulations over a wide range of population sizes show mutation load is only modestly affected by Ne. The forward in time population genetic simulations with SLiM 3 (115) assumed a constant population size, ranging from 200 to 25,000 individuals. We simulated a total of 1,000 evenly distributed exons over a genetic length of 10 cM, with a nonsynonymous to synonymous ratio of 2:1, and a mutation rate of 1e-5 per exon. The selection coefficient of the nonsynonymous mutations was sampled from a gamma distribution as estimated in Huber et al. (98), and synonymous mutations were assumed to be neutral. We considered either additive (h = 0.5; red) or recessive (h = 0; blue) deleterious mutations. We simulated 100,000 generations and recorded genetic diversity and mean fitness of individuals in the population at the end of each run. (A) The number of segregating synonymous sites is proportional to the population size, as expected from neutral theory. (B) Above a long-term population size of 5,000, the population mean fitness is close to the maximum, and independent of population size. Only with a fairly small population size of fewer than 1,000 individuals over prolonged periods of time does fitness become severely reduced due to the fixation of slightly deleterious mutations. This result, i.e., that only a few hundred individuals are necessary to prevent a decline in population fitness and viability, holds true under more realistic simulations of mammalian genomic parameters (33), and when extending the model to allow for beneficial mutations and epistasis (116).