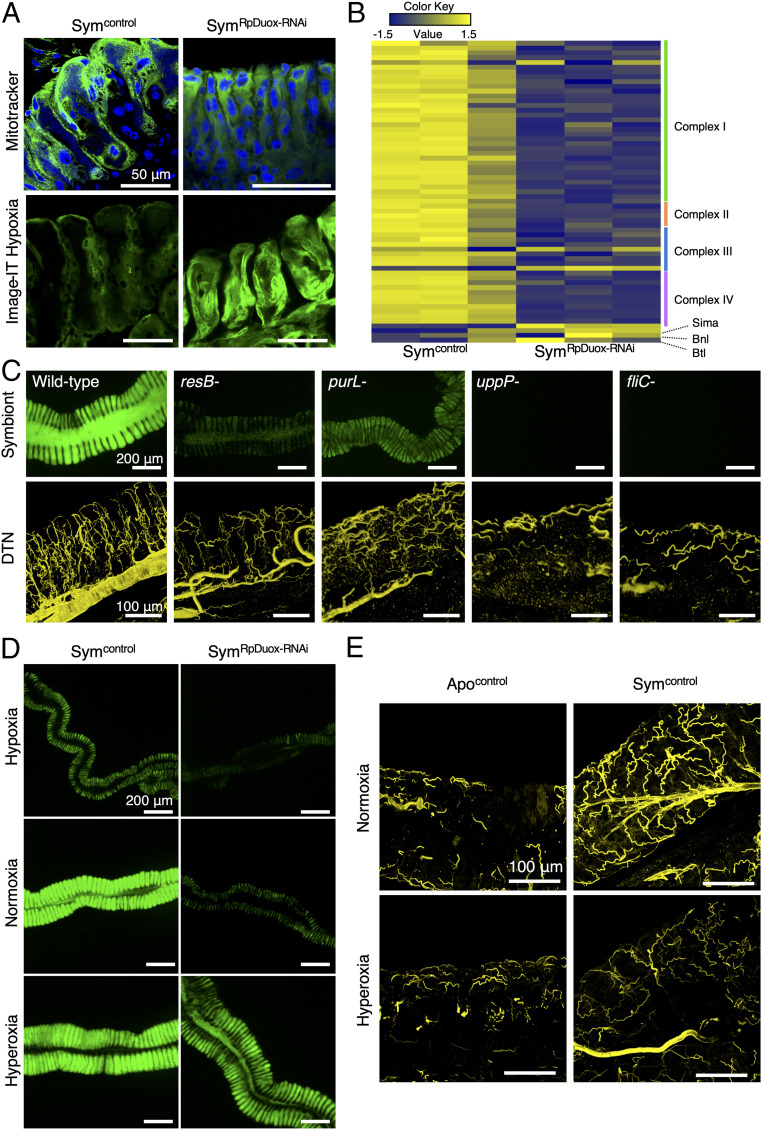
Fig. 4.
Destabilization of the respiratory network by RpDuox-RNAi causes hypoxia in the midgut. (A) Active mitochondria in Sym (Symcontrol) and RpDuox-RNAi (SymRpDuox-RNAi) insects stained by MitoTracker, a membrane potential fluorescence dye, which stains active mitochondria in live cells. Green, MitoTracker; blue, host nuclei (Top). Hypoxia in the M4 revealed by staining with Image-IT hypoxia fluorescence dye, a real-time oxygen detector (green; Bottom). (B) Heat map of relative expression levels of genes related to the host respiratory chain, Sima, Branchless (bnl), and Breathless (btl), in Symcontrol and SymRpDuox-RNAi insects. Each column is a replicate experiment. (C) Colonization level of the wild type, resB-, purL-, uppP-, and fliC-deficient B. insecticola mutants in the symbiotic organ (Top). The DTN of the tracheal network from the symbiotic organs infected with wild type and Burkholderia mutants (Lower). Yellow color indicates the DTN signal of the tracheal network. (D) Observation by epifluorescence microscopy of the midgut of Symcontrol and SymRpDuox-RNAi insects reared in hypoxia (6 to 12% O2), normoxia (21% O2), and hyperoxia (39 to 42% O2) environments. Green is the GFP signal derived from Burkholderia gut symbionts. (E) Tracheal network in the M4 of Apocontrol and Symcontrol insects reared under normoxia and hyperoxia conditions.