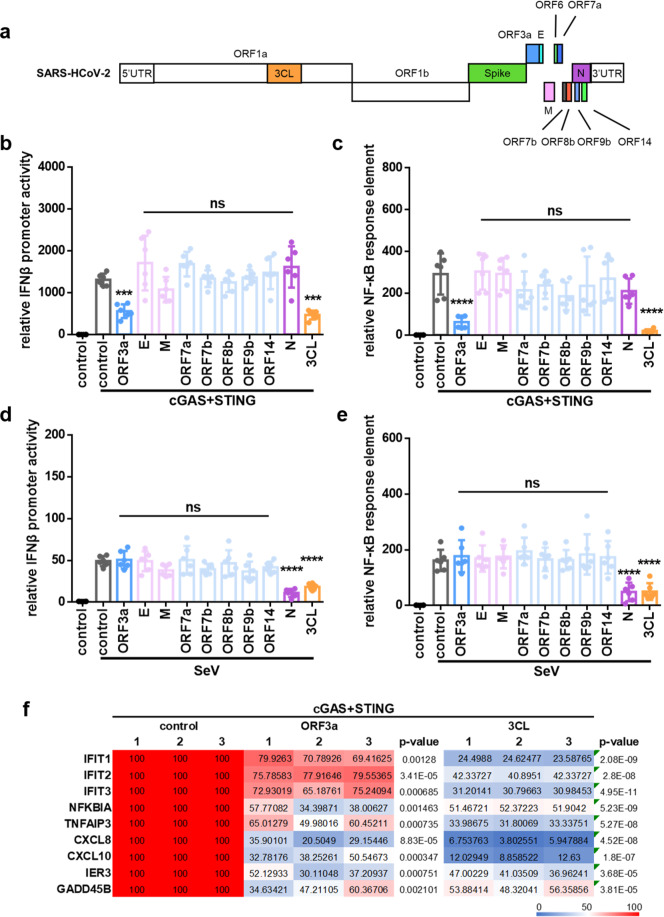
Fig. 1.
Identification of SARS-CoV-2 proteins involved in innate immune suppression. a Schematic diagram of the genomic organization and encoded proteins of SARS-2019 Wuhan-Hu-1 strain (MN908947.3). b, c cGAS-STING-stimulated IFNβ promoter (b) and NF-κB response element (c) activation is inhibited by ORF3a and 3CL. HEK293T cells were co-transfected with IFNβ-Luc (b), NF-κB-Luc (c), and the cGAS and STING expression vectors in the presence or absence of vectors expressing various SARS-CoV-2 proteins as indicated. d, e SeV-induced IFNβ promoter (d) and NF-κB response element (e) activation is inhibited by protein N and 3CL. HEK293T cells were co-transfected with IFNβ-Luc (d) or NF-κB-Luc (e) in the presence or absence of vectors expressing various SARS-CoV-2 proteins as indicated. SeV was added to the culture medium 12 h after transfection. pRL-TK Renilla was used as an internal control (b–e). Transactivation of the luciferase reporter was determined 24 h after transfection (b, c) or 12h after SeV infection (d, e) (n = 6 independent biological experiments). Means and standard deviations are presented. Statistical significance was determined by two-sided unpaired t test, ***p < 0.001; ****p < 0.0001; NS, no significance (b–e). f Inhibition of cGAS-STING-stimulated human genes by ORF3a and 3CL of SARS-CoV-2. HEK293T cells were co-transfected with the cGAS-STING expression vectors in the presence or absence of the ORF3a or 3CL expression vectors as indicated. Human genes activated by cGAS-STING alone served as a positive control and were set to 100%. Total RNA was prepared from HEK293T cells 24 h after transfection and analyzed for the transcriptional level of the indicated genes by RT-qPCR (n = 3 independent biological experiments). Statistical significance was determined by two-sided unpaired t test