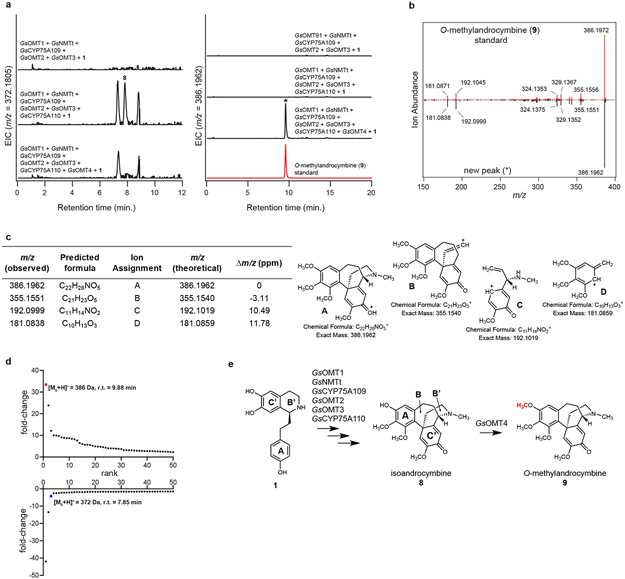
Extended Data Figure 7. Characterization of GsOMT4.
a) Addition of of GsOMT4 into the N. benthamiana transient co-expression system with co-infiltrated 1 leads to consumption of 8 (m/z 372.1805) and production of a new compound corresponding to a methylation (m/z 386.1962), as shown here via LC-MS chromatograms. Comparison to an O-methylandrocymbine (9) standard purified from Colchicum autumnale plants supports the identity of this compound as 9. EIC = extracted ion chromatogram. This result was confirmed in >3 independent experiments. b) MS/MS fragmentation spectrum of the generated m/z 386.1962 product (*) compared to the purified 9 standard, with both compounds fragmented at a collision energy of 20V. This was performed twice, with similar results observed each time. c) Tabulated list and putative structures for ion fragments generated from MS/MS analysis of the m/z 386.1962 product. d) Untargeted metabolite analysis (XCMS) comparing the presence vs. absence of GsOMT4 within the transient co-expression system (n=6 independent replicates for each experimental condition). Shown are unique mass signatures (P < 0.1 between samples, as determined via XCMS) in ranked order based upon their increasing (top panel) or decreasing (bottom panel) fold-change in abundance between the two compared conditions. The mass signature for the product (9, m/z 386.1962) is highlighted in red, while the mass signature of the presumed substrate (m/z 372.1805) is highlighted in blue. r.t. = retention time. e) Proposed reaction catalyzed by GsOMT4, as supported by MS/MS fragmentation, prior labeling studies, and comparison to an isolated 9 standard.