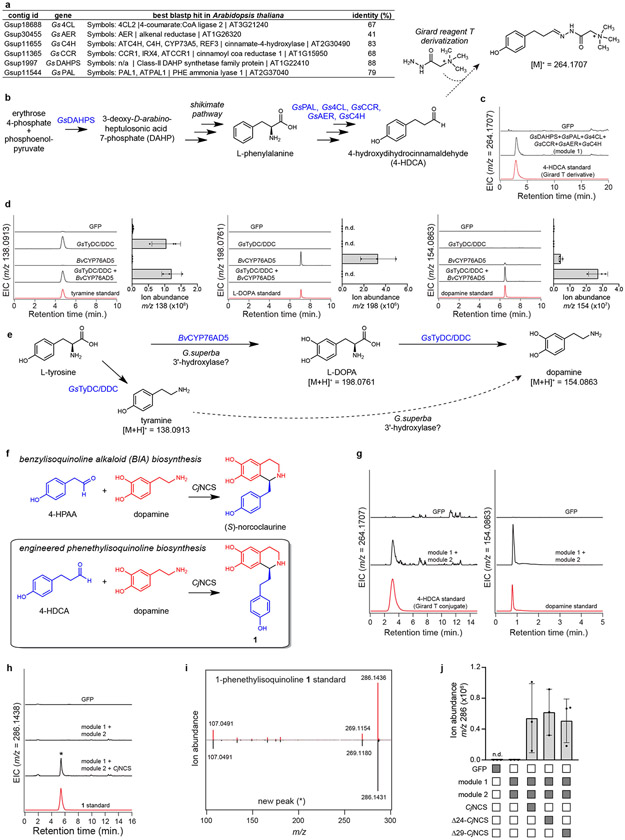
Extended Data Figure 10. Engineering early metabolites in colchicine biosynthesis.
a) List of module 1 biosynthetic genes and their best BLASTP hit in Arabidopsis thaliana. Note that all genes except for GsAER seem to have an ortholog in Arabidopsis with >60% identity, suggesting functional equivalence. b) Generalized pathway for the proposed engineered production of 4-HDCA in N. benthamiana. c) LC-MS chromatograms demonstrating that co-expression of module 1 in N. benthamiana leads to production of 4-HDCA, which was detected as the Girard reagent T derivative (m/z 264.1707). EIC = extracted ion chromatogram. Production of 4-HDCA with module 1 genes was demonstrated three times with similar results each time. d) LC-MS chromatograms (via HILIC analysis) assessing the production of tyramine (left panel, m/z 138.0913), L-DOPA (middle panel, m/z 198.0761), and dopamine (right panel, m/z 154.0863) upon individual and co-expression of GsTyDC/DDC and BvCYP76AD5 (module 2). Next to each set of chromatograms is the corresponding relative quantifications of tyramine (m/z 138), L-DOPA (m/z 198), and dopamine (m/z 154) within each reaction. Shown for each reaction is the mean of 3 distinct biological replicates along with the corresponding standard deviation. n.d. = not detected. Module 2 activity was confirmed in >3 times individual experiments. e) Proposed scheme for the engineered biosynthesis of dopamine. f) Comparison of native CjNCS function within benzylisoquinoline alkaloid (BIA) biosynthesis to the putative reaction required within colchicine alkaloid biosynthesis. g) Co-expression of both module 1 and module 2 genes in N. benthamiana leads to concurrent production of the requisite aldehyde (m/z 264.1707, Girard T derivative), as well as dopamine (m/z 154.0863), as shown here via LC-MS chromatograms. Note that dopamine is observed here with C18 chromatography. Co-expression of both modules was performed >3 times, with similar results each time. h) LC-MS chromatograms for the co-expression of module 1 and module 2 with CjNCS with comparison to an authentic 1 standard. These experiments were performed >3 times, with similar results observed each time. i) MS/MS fragmentation comparison between the newly identified m/z 286.1438 peak (*) and the 1 standard. Both were analyzed with a collision energy of 10V. This MS/MS comparison was performed twice. j) Comparison of wild-type, full-length CjNCS function to N-terminal truncations of 24 (Δ24-CjNCS) and 29 (Δ29-CjNCS) amino acids. Filled-in boxes (gray) indicate the presence of a gene within the co-expression experiment, while an empty box (white) indicates its absence. Shown for each reaction is the mean of 3 biological replicates along with the corresponding standard deviation.