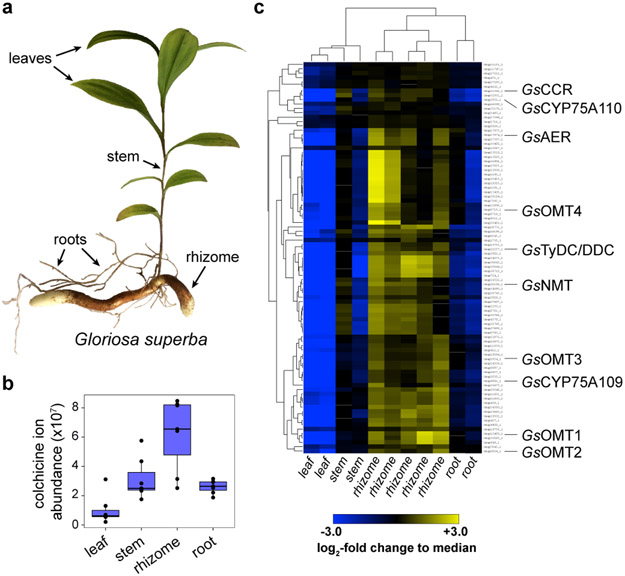
Figure 3. Combined transcriptomics and metabolomics identify notable co-expression of colchicine biosynthetic genes within Gloriosa superba.
a) Tissues from G. superba plants (leaf, stem, rhizome, and root) were used to quantify colchicine alkaloid accumulation and to isolate RNA for subsequent RNA-seq analysis. b) Colchicine accumulates in all tissues, but to the highest level in the rhizome, suggesting this to be the most active site of biosynthesis. Shown is the extracted ion abundance of colchicine, m/z 400.1755 ± 20 ppm. n=7 independent biological replicates for each tissue type, shown here via box and whisker plots. The center line indicates the median; box limits indicate upper and lower quartiles; whiskers indicate 1.5x interquartile range; points outside of the whiskers indicate outliers. c) Hierarchical clustering analysis (distance metric: uncentered Pearson correlation) was performed on contigs with a TMM-normalized, counts per million (CPM) value greater than 25 (for a total of 11,315 out of 38,466 total contigs compared across 11 tissue samples). This analysis identifies an 89 contig cluster that is populated with a substantial number of colchicine biosynthetic genes, as shown here, indicating a high level of co-expression.