Figure 2. MglA-SspA mediated-PigR independent transcription and general binding mode of SspA proteins for RNAPσ70.
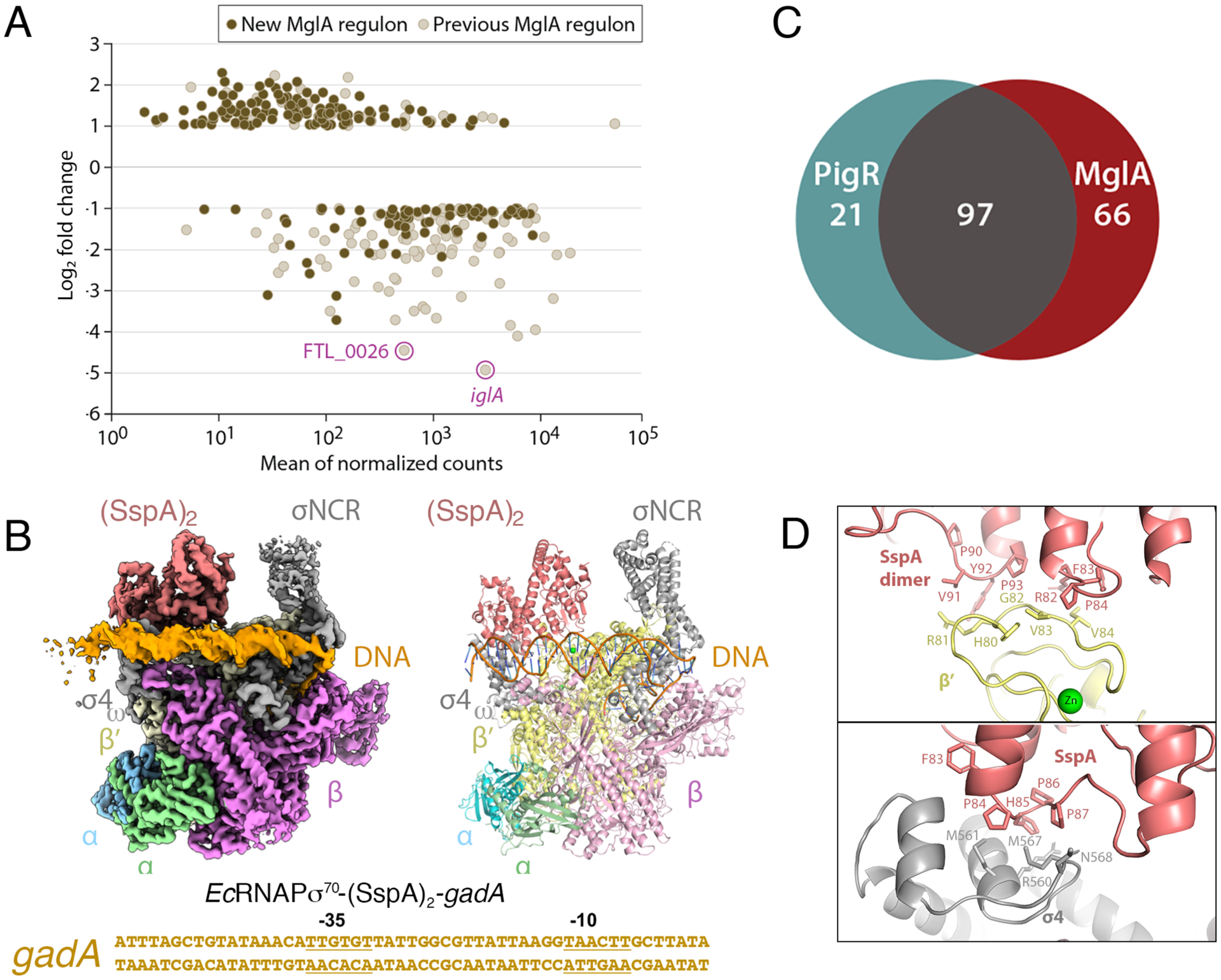
A DESeq2 was used to conduct differential gene expression analysis between WT and ΔmglA mutant RNA-Seq libraries. The graph shows log2 fold change in transcript abundance in ΔmglA compared to WT cells. Previously described MglA-controlled transcripts (such as iglA and FTL_0026) are in beige (with the iglA and FTL_0026 transcripts highlighted). Newly identified MglA-transcripts are in brown. B Venn diagram indicating MglA-regulated genes that are controlled by PigR and those that are not. Most genes regulated by PigR are also regulated by MglA-SspA but MglA-SspA regulates multiple genes independently of PigR. C Cryo-EM EcRNAPσ70-(SspA)2-gadA DNA structure. Left is cryo-EM map, right is ribbon diagram and below is the sequence of the gadA promoter used in the structure, with −35 and −10 elements labeled. The complex is shown in the same orientation as Figure 1A. D Close up views of contact points between EcSspA and EcRNAPσ70.
