Abstract
The platelet endothelial aggregation receptor-1 (PEAR1) rs12041331 variant has been identified as a genetic determinant of platelet aggregation in response to antiplatelet therapies, including aspirin. However, association with atherothrombotic cardiovascular events is less clear, with limited evidence from large trials. Here, we tested association of rs12041331 with cardiovascular events and aspirin use in a randomized trial population of healthy older individuals. We undertook post hoc analysis of 13,547 participants of the ASPirin in Reducing Events in the Elderly (ASPREE) trial, median age 74 years. Participants had no previous diagnosis of atherothrombotic cardiovascular disease at enrollment, and were randomized to either 100 mg daily low-dose aspirin or placebo for median 4.7 years follow-up. We used Cox proportional hazard regression to model the relationship between rs12041331 and the ASPREE primary cardiovascular disease (CVD) end point, and composites of major adverse cardiovascular events (MACE) and ischemic stroke (STROKE); and bleeding events; major hemorrhage (MHEM) and intracranial bleeding (ICB). We performed whole-population analysis using additive and dominant inheritance models, then stratified by treatment group. Interaction effects between genotypes and treatment group were examined. We observed no statistically significant association (P < 0.05) in the population, or by treatment group, between rs12041331 and cardiovascular or bleeding events in either model. We also found no significant interaction effects between rs12041331-A and treatment group, for CVD (P = 0.65), MACE (P = 0.32), STROKE (P = 0.56), MHEM (P = 0.59), or ICB (P = 0.56). The genetic variant PEAR1 rs12041331 is not associated with cardiovascular events in response to low-dose aspirin in a healthy elderly population.
Interindividual variability in response to antiplatelet medications, including aspirin, can result in inadequate platelet inhibition and subsequent cardiovascular and cerebrovascular events. Clinical and anthropometric factors are known to contribute to variable aspirin efficacy, including age, sex, body mass index (BMI), diabetes mellitus (DM), compliance, and certain drug-drug interactions.1–3 In addition, heritability estimates, as assessed by ex vivo agonist-stimulated platelet aggregation, suggest that a large proportion of variation in response to aspirin may be attributable to genetic factors.4 Indeed, multiple pharmacogenetic investigations ranging from candidate gene reports to genomewide association studies have been conducted to identify novel genetic determinants of aspirin response. Identification of genetic polymorphisms that lead to suboptimal platelet control may have important implications in primary and secondary prevention of adverse clinical events in patients with an indication for this medication.
The platelet endothelial aggregation receptor 1 (PEAR1) gene encodes a type 1 membrane protein that is highly expressed in platelets and endothelial cells5 and critical in hematological processes, including megakaryopoiesis, thrombopoiesis, and aαIIbβ3-mediated platelet aggregation.6,7 Genetic variants in the PEAR1 gene have been associated with platelet aggregation8,9 and in response to antiplatelet agents, including aspirin,10–12 clopidogrel,13 ticagrelor,14 and prasugrel.15 Most notably, an intronic variant rs12041331 in the PEAR1 gene is among the strongest genetic modifiers of platelet aggregation, particularly in those treated with aspirin, and results in allele-specific differences in H3K4Me1 methylation leading to differential gene and protein expression.11,16,17 Despite the evidence demonstrating the influence of PEAR1 rs12041331 on agonist-stimulated platelet aggregation in aspirin-treated patients, the impact of this variant on thrombotic and bleeding events remains unclear.
In this investigation, we assessed the interaction between PEAR1 rs12041331 and randomized aspirin use in 13,547 participants of the Aspirin in Reducing Events in the Elderly (ASPREE) trial. Specifically, individuals with no previous diagnosis of atherothrombotic cardiovascular disease (CVD) were randomized to either 100 mg daily low-dose aspirin or placebo for median 4.7 years and relationships between rs12041331 genotypes and cardiovascular events, as well as clinically significant bleeding events, were examined. Moreover, gene x environment (i.e., aspirin treatment) interaction models were used to test whether the effect of rs12041331 genotypes on clinical end points varied based on aspirin use.
METHODS
Study population
Details of the ASPREE trial are described previously.18,19 Briefly, ASPREE was a double-blind, randomized (1:1), placebo-controlled trial to examine the effect of daily low-dose aspirin on disability-free survival in healthy older individuals.19–21 At enrollment, the 19,114 participants of the ASPREE trial had no current symptoms or history of cardiovascular events, including myocardial infarction, heart failure, stroke, transient ischemic attack, atrial fibrillation, or high blood pressure. Participants also had no dementia diagnosis, physical disability, or illness likely to cause death within 5 years at enrollment. Participants were followed prospectively, and monitored for a range of primary and secondary outcomes, including cardiovascular events, dementia, persistent disability, and cancer. Participants were enrolled from March 2010 through December 2014, with follow-up data collected until June 2017 (median follow-up of 4.7 years). All investigational protocols were approved by local institutional review boards across Australia and the United States and conformed to the principles outlined in the Declaration of Helsinki. Each participant gave informed consent prior to enrollment.
Outcomes
We analyzed the relationship between aspirin use and the common variant PEAR1 rs12041331, in relation to clinically adjudicated cardiovascular and bleeding events in ASPREE. End points analyzed included the primary CVD end point in the ASPREE study, and composites of major cardiovascular events considered most likely to be impacted by aspirin; including major adverse cardiovascular events (MACEs) and ischemic stroke (STROKE); and clinically significant bleeding events, including major hemorrhage (MHEM) and intracranial bleeding (ICB). Details of ASPREE end points have been described previously.21
Genetic analysis
Ethical approval for genetic analysis was obtained from the Alfred Hospital Human Research Ethics Committee (390/15). Genomewide single-nucleotide polymorphism (SNP) genotyping of 14,177 samples collected by the ASPREE Healthy Ageing Biobank was performed using the Axiom 2.0 Precision Medicine Diversity Research Array (Thermo Fisher Scientific, Chino, CA) following standard protocols, at the Ramaciotti Centre for Genomics, University of New South Wales, Australia. Samples were excluded based on poor quality control metrics or genotyping performance (N = 426) and other filters, including sex discordance (N = 80), and relatedness (N = 124). In the final dataset, a total of 13,547 samples from individuals of predominantly non-Finish European ancestry were available for genetic analyses. Individual variants were excluded based on genotyping rate (< 95%) and Hardy-Weinberg equilibrium. Imputation was performed on European-ancestry samples using the haplotype reference consortium panel on the University of Michigan imputation server.22 Post-imputation quality control removed all variants with < 0.3 imputation quality scores. The imputation quality score of the variant rs12041331 was R2 = 0.99.
Statistical analysis
We used Cox proportional hazards regression models to analyze the relationship between rs12041331-A allele and cardiovascular events, adjusting for variables related to cardiovascular risk, including age, sex, smoking status, alcohol intake, high density lipoproteins (HDL), low density lipoprotein (LDL)-cholesterol, triglycerides (TGs), total cholesterol (TC), hypertension, BMI, DM, and statin use prior to enrollment. Analysis of the entire cohort (N = 13,547) was performed using an additive model of inheritance (GG vs. AG vs. AA) and dominant model (GG vs. AG/AA), followed by stratified analysis of the placebo (N = 6,806) and aspirin (N = 6,741) groups separately. Then, the modifying effect of the rs12041331-A genotype on association between incident CVD events and aspirin treatment was analyzed using an interaction term in a multivariable Cox regression model using participants from both groups, under both the additive and dominant genetic models. We calculated the minimal detectable effect for all end points under both models. All analyses were performed with Stata 15.1 (Stata, College Station, TX) and R (R Core Team, 2014). P values < 0.05 were considered as statistically significant (two-sided). Figures were produced using the package ggplot2.
RESULTS
The 13,547 genotyped participants had a median age of 73.9 years and were 54.7% women. The distributions ofage, sex, plasma lipids, and BMI variables were balanced between the groups (Table 1). Genotype distribution for rs12041331 was also balanced between the placebo and aspirin groups, with GA heterozygotes found at 16.3% (N = 1,112) and 15.3% (N = 1,034), respectively, and AA homozygotes at 0.9% (N = 64) and 0.9% (N = 60), respectively (Figure 1, Table 2).
Table 1. Characteristics of genotyped participants by treatment status.
| Placebo N = 6,806 |
Aspirin N = 6,741 |
Total N = 13,547 |
|
|---|---|---|---|
| Age at randomization, years | 73.9 (71.7, 77.3) | 73.9 (71.7, 77.3) | 73.9 (71.7, 77.3) |
| 65–73.9 years | 3,485 (51.2%) | 3,446 (51.1%) | 6,931 (51.2%) |
| ≥74 years | 3,321 (48.8%) | 3,295 (48.9%) | 6,616 (48.8%) |
| Sex | |||
| Female | 3,716 (54.6%) | 3,691 (54.8%) | 7,407 (54.7%) |
| Male | 3,090 (45.4%) | 3,050 (45.2%) | 6,140 (45.3%) |
| Blood lipid levels, mg/dl at baseline | |||
| LDL-C | 119.9 (96.7, 140.0) | 116.0 (96.0, 139.2) | 116.0 (96.7 to 139.2) |
| HDL-C | 58.0 (50.3, 69.6) | 58.0 (50.0, 69.6) | 58.0 (50.3 to 69.6) |
| TG | 106.3 (79.7, 141.7) | 106.3 (79.7, 141.7) | 106.3 (79.7 to 141.7) |
| TC | 201.1 (177.9, 228.2) | 201.1 (177.9, 228.2) | 201.1 (177.9 to 228.2) |
| BMI kg/m2, (baseline) | 27.4 (24.9–30.5) | 27.4 (24.9–30.5) | 27.4 (24.9 to 30.5) |
| Underweight, < 20 | 112 (1.7%) | 119 (1.8%) | 231 (1.7%) |
| Normal, 20–24.9 | 1,635 (24.1%) | 1,643 (24.5%) | 3,278 (24.3%) |
| Overweight, 25–29.9 | 3,112 (45.9%) | 3,037 (45.2%) | 6,149 (45.6%) |
| Obese, ≥ 30 | 1,917 (28.3%) | 1,914 (28.5%) | 3,831 (28.4%) |
| Statin usea | 2,369 (34.8%) | 2,384 (35.4%) | 4,753 (35.1%) |
| Hypertensiona | 5,056 (74.3%) | 4,928 (73.1%) | 9,984 (73.7%) |
Data summaries are median (interquartile range) for continuous measures, and n (%) for categorical measures.
BMI, body mass index; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride.
Self-reported.
Figure 1.

Schematic overview (graphical abstract). We examined the relationship between rs12041331 genotypes and cardiovascular and bleeding events in a randomized placebo-controlled study population of 13,547 participants of the ASPirin in Reducing Events in the Elderly (ASPREE) trial. Participants had no previous diagnosis of atherothrombotic cardiovascular disease (CVD) at enrollment, and were randomized to either 100 mg daily low-dose aspirin or placebo for a median 4.7 years of follow-up. End points analyzed included the primary CVD end point in the ASPREE study, and composites of major adverse cardiovascular events (MACE) and ischemic stroke; and bleeding events, including major hemorrhage and intracranial bleeding. Details of ASPREE end points are described previously.21 The modifying effect of the rs12041331-A genotype on association with cardiovascular and bleeding events and aspirin treatment was analyzed using an interaction term in a multivariable Cox regression model.
Table 2. Genotype distribution by treatment status.
| rs12041331 | Placebo | Aspirin | Total |
|---|---|---|---|
| GG | 5,630 (82.7%) | 5,647 (83.8%) | 11,277 (83.2%) |
| GA | 1,112 (16.3%) | 1,034 (15.3%) | 2,146 (15.8%) |
| AA | 64 (0.9%) | 60 (0.9%) | 124 (0.9%) |
| 6,806 | 6,741 | 13,547 |
Genomewide single-nucleotide polymorphism genotyping of 14,177 samples collected by the ASPirin in Reducing Events in the Elderly (ASPREE) Healthy Ageing Biobank was performed using the Axiom 2.0 Precision Medicine Diversity Research Array (Thermo Fisher Scientific) following standard protocols. A total of 13,547 samples passed post-genotyping quality control (QC) filters. Samples were excluded based on filters related to sex discordance, genotyping rate (< 95%), relatedness, Hardy–Weinberg equilibrium, and ethnicity. Imputation was performed on European-ancestry samples using the haplotype reference consortium panel on the Michigan imputation server.22 Post-imputation QC removed all variants with < 0.3 imputation quality scores. The imputation quality score of the variant rs12041331 was R2 = 0.99.
We did not observe evidence of association between PEAR1 rs12041331 and cardiovascular or bleeding events when stratifying the cohort by treatment effect (Table 3), based on using an additive (Figure 2) or dominant (Figure 3) models of inheritance. In the placebo group, when comparing major allele homozygotes to heterozygotes or minor allele homozygotes using the additive model, no difference in event rate was noted for CVD (P = 0.27 and P = 0.62, respectively), MACE (P = 0.66 and P = 0.73, respectively), STROKE (P = 0.92 and P = 0.95, respectively), MHEM (P = 0.57 and P = 0.61, respectively) or ICB (P = 0.92 and P = 0.95, respectively). Hazard ratio (HR) point estimates (Table 3) were generally in the same direction as observed when evaluating the entire cohort (see below; Table S1).
Table 3. Association of rs12041331 genotypes with ASPREE CVD and bleeding events in aspirin and placebo treatment groups.
| ASPREE CVD end point | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Placebo | Aspirin | ||||||||
| No CVD n (%) |
CVD n (%) |
HR adj (95% CI) | P value | No CVD n (%) |
CVD n (%) |
HR adj (95% CI) | P value | ||
| GG | 5,351 (95.0) | 279 (5.0) | 1 | - | GG | 5,406 (95.7) | 241 (4.3) | 1 | - |
| GA | 1,065 (95.8) | 47 (4.2) | 0.84 (0.61, 1.15) | 0.27 | GA | 1,001 (96.8) | 33 (3.2) | 0.76 (0.53, 1.09) | 0.14 |
| AA | 60 (93.8) | 4 (6.3) | 1.28 (0.48, 3.40) | 0.62 | AA | 58 (96.7) | 2 (3.3) | 0.82 (0.21, 3.21) | 0.78 |
| MACE | |||||||||
| Placebo | Aspirin | ||||||||
|
No MACE n (%) |
MACE n (%) |
HR adj (95% CI) | P value |
No MACE n (%) |
MACE n (%) |
HR adj (95% CI) | P value | ||
| GG | 5,418 (96.2) | 212 (3.8) | 1 | - | GG | 5,469 (96.8) | 178 (3.2) | 1 | - |
| GA | 1,073 (96.5) | 39 (3.5) | 0.93 (0.66, 1.31) | 0.66 | GA | 1,011 (97.8) | 23 (2.2) | 0.70 (0.46, 1.09) | 0.11 |
| AA | 61 (95.3) | 4 (6.3) | 1.22 (0.40, 3.71) | 0.73 | AA | 58 (96.7) | 2 (3.3) | 1.15 (0.29, 4.54) | 0.85 |
| STROKE | |||||||||
| Placebo | Aspirin | ||||||||
|
No STROKE n (%) |
STROKE n (%) |
HR adj (95% CI) | P value |
No STROKE n (%) |
STROKE n (%) |
HR adj (95% CI) | P value | ||
| GG | 5,534 (98.3) | 96 (1.7) | 1 | - | GG | 5,773 (98.7) | 74 (1.3) | 1 | - |
| GA | 1,093 (98.3) | 19 (1.7) | 1.02 (0.63, 1.68) | 0.92 | GA | 1,023 (98.9) | 11 (1.1) | 0.81 (0.43, 1.52) | 0.51 |
| AA | 0.94 (0.13, 6.63) | AA | 1.36 (0.19, 9.83) | ||||||
| MHEM | |||||||||
| Placebo | Aspirin | ||||||||
|
No MHEM n (%) |
MHEM n (%) |
HR adj (95% CI) | P value |
No MHEM n (%) |
MHEM n (%) |
HR adj (95% CI) | P value | ||
| GG | 5,479 (97.3) | 96 (1.7) | 1 | GG | 5,448 (96.5) | 199 (3.5) | 1 | - | |
| GA | 1,093 (98.3) | 19 (1.7) | 0.88 (0.57, 1.34) | 0.57 | GA | 998 (96.5) | 36 (3.5) | 1.02 (0.71, 1.45) | 0.92 |
| AA | 63 (98.4) | 1 (1.6) | 0.60 (0.08, 4.25) | 0.61 | AA | 60 (100.0) | 0 (0) | NA | - |
| ICB | |||||||||
| Placebo | Aspirin | ||||||||
|
No ICB n (%) |
ICB n (%) |
HR adj (95% CI) | P value |
No ICB n (%) |
ICB n (%) |
HR adj (95% CI) | P value | ||
| GG | 5,594 (99.4) | 36 (0.6) | 1 | - | GG | 5,585 (98.9) | 62 (1.1) | 1 | - |
| GA | 1,106 (99.5) | 6 (0.5) | 1.03 (0.63, 1.68) | 0.92 | GA | 1,026 (99.2) | 8 (0.8) | 0.81 (0.43, 1.52) | 0.51 |
| AA | 63 (98.4) | 1 (1.6) | 0.94 (0.13, 6.63) | 0.95 | AA | 60 (100.0) | 0 (0) | - | - |
We used Cox proportional hazard regression to model the relationship between rs12041331 genotypes with cardiovascular and bleeding events. End points analyzed included the ASPREE CVD end point, and subcomponents of MACE and STROKE; and the ASPREE clinically significant bleeding end point, and subcomponents of MHEM and ICB. Details of ASPREE end points are described previously.21
ASPREE, ASPirin in Reducing Events in the Elderly; CI, confidence interval; CVD, cardiovascular disease; HR, hazard ratio; ICB, intracranial bleeding; MACE, major adverse cardiovascular event; MHEM, major hemorrhage; NA, not applicable; STROKE, ischemia stroke.
HR adj: hazard ratio, adjusted for age, sex, smoking status, alcohol intake, high density lipoproteins, low density lipoprotein cholesterol, triglycerides, total cholesterol, hypertension, body mass index, diabetes mellitus, and statin use prior to enrolment (multivariable Cox regression analysis).
Figure 2.

Cumulative incidence curves (additive genetic model). We undertook cumulative incidence analysis of cardiovascular and bleeding events in aspirin and placebo groups, stratified by rs12041331 genotypes using an additive model of genetic inheritance; GG wild type (blue curves), AG heterozygotes (red curves), and AA homozygotes (yellow curves). Events analyzed included the primary cardiovascular disease (CVD) end point in the ASPirin in Reducing Events in the Elderly (ASPREE) study (a, b), and composites of major adverse cardiovascular events (MACE) (c, d) and ischemic stroke (STROKE) (e, f); and the ASPREE clinically significant bleeding end point subcomponents of major hemorrhage (MHEM) (g, h) and intracranial bleeding (ICB) (i, j).
Figure 3.
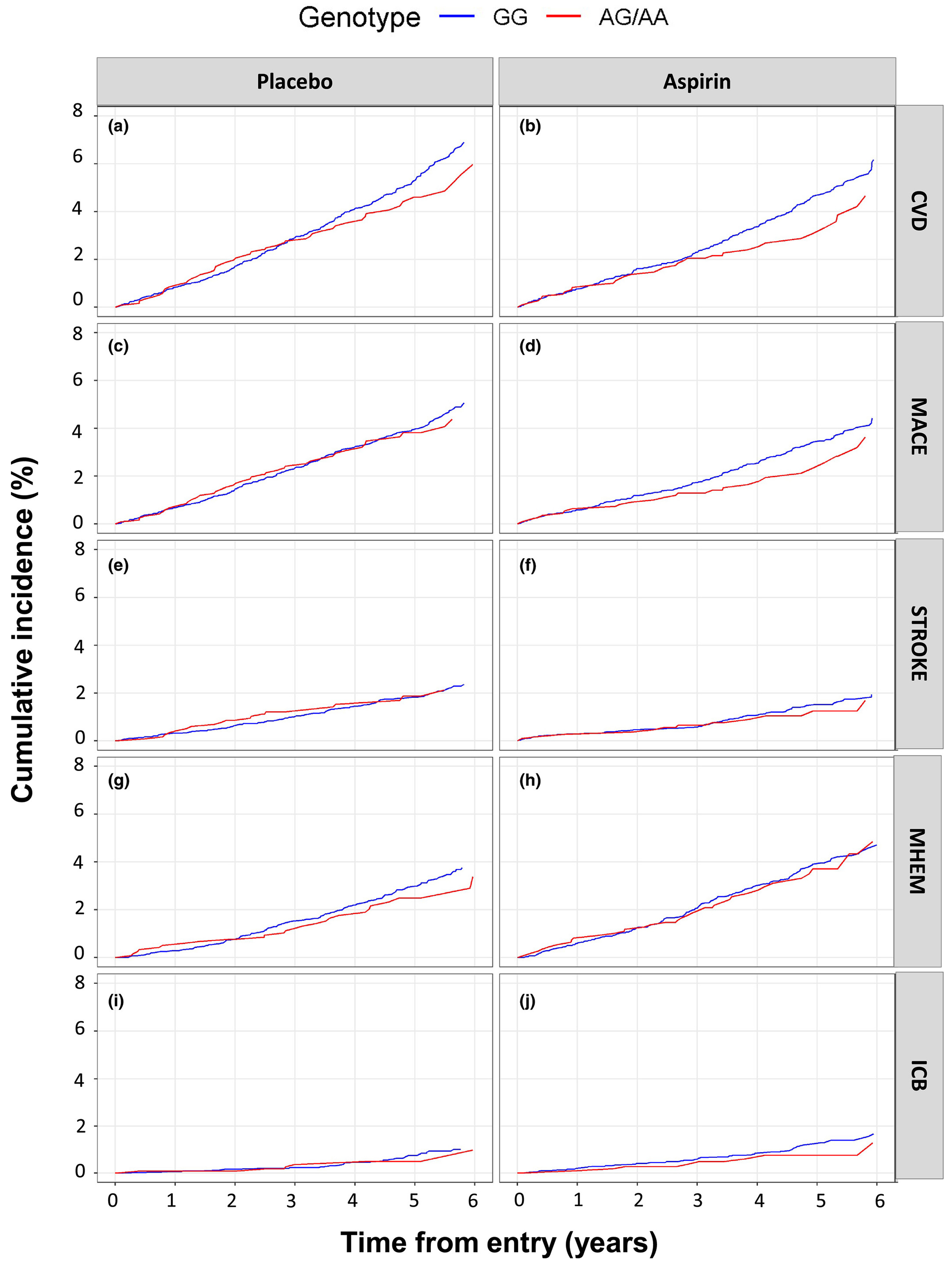
Cumulative incidence curves (dominant genetic model). We undertook cumulative incidence analysis of cardiovascular and bleeding events in aspirin and placebo groups, stratified by rs12041331 genotypes using a dominant model of genetic inheritance; GG wild type (blue curves) vs. AG/AA carriers (red curves). Events analyzed included the primary cardiovascular disease (CVD) end point in the ASPirin in Reducing Events in the Elderly (ASPREE) study (a, b), and composites of major adverse cardiovascular events (MACEs) (c, d) and ischemic stroke (STROKE) (e, f); and the ASPREE clinically significant bleeding end point subcomponents of major hemorrhage (MHEM) (g, h) and intracranial bleeding (ICB) (I, j).
Similarly, there was no evidence to suggest that PEAR1 rs12041331 genotype impacted cardiovascular or bleeding event rate in participants treated with aspirin (Table 3). In individuals randomized to aspirin, rs12041331 A-allele carrier status did not result in statistically significant differences in atherothrombotic event rates (CVD, MACE, and STROKE) or bleeding events (MHEM and ICB) regardless if individuals carried one (CVD P = 0.14, MACE P = 0.11, STROKE P = 0.51, MHEM P = 0.92, and ICB P = 0.51) or two (CVD P = 0.78, MACE P = 0.85, STROKE P = 0.76, MHEM P = N/A, and ICB P = 0.76) copies of the A-allele (Table 3). We repeated these analyses using a dominant model of inheritance (wild-type GG vs. all AA/AG carriers) and also found no significant associations (Table S2).
In the combined cohort (both aspirin-treated and placebo-treated groups), no association was observed between rs12041331 A-allele carrier status and atherothrombotic events for either the additive (Table S1) or dominant (Table S2) genetic models. Specifically, no statistical difference in event rates for CVD, MACE, and STROKE were seen when comparing PEAR1 rs12041331 major allele homozygotes to either heterozygotes (HR = 0.81, 95% confidence interval (CI) 0.64–1.02, P = 0.07; HR = 0.84, 95% CI 0.64–1.09, P = 0.19; and HR = 0.94, 95% CI 0.64–1.38, P = 0.74, respectively) or minor allele homozygotes (HR = 1.10, 95% CI 0.50–2.44, P = 0.50; HR = 1.22, 95% CI 0.51–2.90, P = 0.65; and HR = 1.13, 95% CI 0.28–4.53, P = 0.86, respectively). Similarly, no correlation between genotype and MHEM or ICB was observed when comparing rs12041331 major allele homozygotes to either heterozygotes (HR = 0.96, 95% CI 0.73–1.26, P = 0.74 and HR = 0.94, 95% CI 0.64–1.38, P = 0.74, respectively) or minor allele homozygotes (HR = 0.27, 95% CI 0.04–1.93, P = 0.19 and HR = 1.13, 95% CI 0.28–4.53, P = 0.28, respectively).
Using a multivariable Cox regression model, adjusting for age, sex, smoking status, alcohol intake, HDL, LDL-cholesterol, TG, TC, hypertension, BMI, DM, and statin use prior to enrollment, we found no evidence of an interaction effect between rs12041331 genotypes and aspirin treatment under the additive or dominant inheritance model, for incident CVD, MACE, ischemic stroke, MHEM, and ICB (Table 4, Tables S3-S8). For calculations of minimal detectable effects, under both models for all end points, see Tables S9 and S10.
Table 4. Tests of interaction effect between treatment group and the rs12041331-A genotype on ASPREE CVD and bleeding events.
| CVD |
MACE |
STROKE |
MHEM |
ICB |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs12041331 | z | P value | z | P value | z | P value | z | P value | z | P value |
| GA # Aspirin | −0.46 | 0.649 | −0.99 | 0.321 | −0.58 | 0.561 | 0.54 | 0.591 | −0.58 | 0.561 |
| AA # Aspirin | −0.052 | 0.606 | −0.09 | 0.928 | 0.25 | 0.802 | −35.2 | – | 0.25 | 0.802 |
The modifying effect of the rs12041331-A genotype on association between incident CVD events and aspirin treatment was analyzed using an interaction term in a multivariable Cox regression model using participants from both groups, adjusted for age, sex, smoking status, alcohol intake, high density lipoproteins, low density lipoprotein cholesterol, triglycerides, total cholesterol, hypertension, body mass index, diabetes mellitus, and statin use prior to enrolment. End points analyzed included the ASPREE CVD end point, and subcomponents of MACE and STROKE; and the ASPREE clinically significant bleeding end point, and subcomponents of MHEM and ICB. Details of ASPREE end points are described previously.21
ASPREE, ASPirin in Reducing Events in the Elderly; CVD, cardiovascular disease; ICB, intracranial bleeding; MACE, major adverse cardiovascular event; MHEM, major hemorrhage; STROKE, ischemia stroke.
DISCUSSION
There is considerable evidence that the PEAR1 receptor is a critical part of platelet aggregation in response to several agonists, and that rs12041331 is a strong genetic determinant of platelet aggregation.5,6,12,23,24 However, there are less data regarding the impact of this variant on cardiovascular event risk in untreated individuals or in patients prescribed antiplatelet medication. In the current study, we sought to evaluate the impact of PEAR1 SNP rs12041331, a well-described genetic variant implicated in aspirin-related platelet function, on MACE and clinically significant bleeding events in a large healthy cohort of elderly individuals randomized to either aspirin or placebo in the ASPREE trial. In analyses of the entire cohort or when stratifying by treatment effect (i.e., aspirin vs. placebo), using either an additive or dominant model of inheritance, we observed no evidence of association for rs12041331 with increased risk of experiencing a thrombotic or bleeding event. Furthermore, we observed no significant interaction effects among PEAR1 genotype, aspirin use, and clinical outcomes using either an additive or dominant model.
Since its identification in 2005, multiple investigations have been published highlighting the role of PEAR1 in several important biological processes.5 The most extensively studied of these processes has been platelet aggregation; however, other proposed functions of this receptor include maintenance of endothelial cell function,13,25 megakaryopoiesis and thrombopoiesis,7 neoangiogenesis,26 leukocyte function,17 and neuronal phagocytosis.27,28 These and other publications have prompted numerous genetic studies attempting to correlate PEAR1 polymorphisms with related phenotypes and diseases. From these investigations, rs12041331 has been among the most thoroughly studied, with the strongest effect on platelet-related phenotypes, and established functional effect on PEAR1 expression. Previously, rs12041331 A-allele carriers have been shown to have significantly lower DNA methylation in a CpG-island, which contains binding sites for the methylation-sensitive transcription factor CTCF compared with GG homozygotes, presumably leading to differences in gene and protein expression observed by others at this locus.11,16
Given the effect of rs12041331 on PEAR1 expression and platelet-related processes, recent investigations have focused on the impact of this polymorphism on cardiovascular-related diseases with mixed results. Initial reports in patients with percutaneous coronary intervention treated with aspirin and clopidogrel showed that A-allele carriers of this variant experienced MACE (HR = 2.62; 95% CI 0.96–7.10, P = 0.06) and cardiovascular-related death (HR = 3.97, 95% CI 1.10–14.31, P = 0.04) more frequently compared with GG homozygotes.29 However, these results were not confirmed in a Flemish population, which reported no association with several different cardiovascular and cerebrovascular phenotypes when using rs12566888 as a proxy SNP for rs12041331 (R2 = 0.99, D = 1.00 with each other).30 These latter findings were consistent with a subsequent publication showing no association between PEAR1 rs12041331 and ischemic events in Chinese patients with an acute coronary syndrome treated with aspirin and clopidogrel (HR = 1.30, 95% CI 0.74–2.29, P = 0.07), although it should be noted that statistical power was likely quite low in this study given a limited sample size of 196 patients.31
Most recently, Xu et al. prospectively assessed rs12041331 in over 2,400 Chinese patients with acute coronary syndrome or stable coronary artery disease undergoing stent implantation and treated with aspirin and clopidogrel.32 Based on PEAR1 genotype, they observed significantly different ADP-induced platelet aggregation (29.1, 25.4, and 22.9 for genotypes GG, GA, and AA, respectively, P = 0.0005) and detected differences in the 30-day incidence of a composite cardiovascular phenotype consisting of cardiovascular death, nonfatal myocardial infarction, and ischemic stroke (HR = 2.78, 95% CI 1.13–6.82, P = 0.03). Positive associations with deep vein thrombosis in those with sticky platelet syndrome as well as coronary artery aneurysm in individuals with Kawasaki disease have also been shown by PEAR1 rs12041331 genotype.33,34
Further studies have suggested a genotype x aspirin interaction exists whereby aspirin-treated carriers ofrs12041331 minor (A) allele have enhanced risk of experiencing a clinical event compared with carriers of the same allele who are not treated with aspirin.29 Specifically, it was observed in patients with stable coronary artery disease of European descent that rs12041331 A-allele carriers treated with aspirin have significantly increased risk of myocardial infarction (odds ratio (OR) = 2.08, 95% CI 1.01–4.09, P = 0.048) compared with those who were not exposed to aspirin (OR = 0.25, 95% CI 0.03–2.03, P = 0.19). Similar trends were observed using a composite cardiovascular outcome consisting of death, nonfatal stroke, and nonfatal myocardial infarction (OR = 1.62, 95% CI 0. 91–2.90, P = 0.10 and OR = 0.54, 95% CI 0.22–1.31, P = 0.16, respectively).29 Evidence also suggests that the PEAR1 receptor may be able to exert cardiovascular-related effects through processes that are independent of platelet function (e.g., endothelial-related effects). Investigation of other molecular processes that are regulated by PEAR1 would likely allow for more targeted investigation of effects that may modify cardiovascular risk.
It is noteworthy that all previous studies discussed above have been undertaken in populations known to be affected by CVD. The present study, however, has been undertaken in a highly selected population of healthy older individuals with no previous diagnosis of atherothrombotic CVD, and in the context of primary prevention. Our study results, therefore, must be interpreted accordingly.
In our investigation, which is the largest study of PEAR1 rs12041331 and cardiovascular outcomes conducted to date, we did not observe significant association between rs12041331 and cardiovascular events or clinically significant bleeding in the entire cohort, regardless of aspirin use (Table 3). We also found no significant effects in interaction analyses using either the additive (Table 4, Tables S3-S7) or dominant (Table S8) genetic model. Given that the minor allele of this variant resulted in lower agonist-stimulated platelet aggregation in prior investigations, we also assessed whether aspirin-dependent effects could modify the relationship between PEAR1 genotype and clinically significant bleeding events. Consistent with our analyses pertaining to thrombotic outcomes, no association was observed between rs12041331 and bleeding in both placebo-randomized or aspirin-randomized individuals and no statistically significant interaction was observed between aspirin treatment and genotype on occurrence of bleeding events.
Although our data suggest that this polymorphism is not an important contributor to clinical events in a healthy older population, in the setting of primary prevention, additional studies are warranted, investigating the effect of rs12041331 on MACE in other populations, and including the general population, and patients with higher cardiovascular burden, such as those following a coronary intervention procedure, and/or on a dual antiplatelet therapy regimen.
Strengths of our study include the sample size, being the largest evaluation of PEAR1 rs12041331 conducted to date, with regard to cardiovascular outcomes and aspirin use. To our knowledge, it is also the only study to examine the effect of rs12041331 on clinically significant bleeding events. In addition, the prospective, randomized nature of the study design implemented in the ASPREE trial allowed us to confidently assess the interaction between PEAR1 genotype and aspirin use on adverse events in cohorts that were well-matched for clinical and anthropometric characteristics known to influence cardiovascular and bleeding risk.18 Central adjudication of cardiovascular and bleeding events allowed for blinded evaluation of suspected outcomes and ultimately allowed us to maximize data consistency while minimizing potential biases.
Limitations of our study include the highly selected nature of the ASPREE population, and the predominance of European ancestry, which limit the generalizability of our results to other populations. Participants enrolled into the ASPREE trial were elderly, generally healthy individuals with no known atherothrombotic CVD and were randomized to aspirin in the context of primary prevention. Therefore, our study results may not be indicative of data obtained in patients with more severe cardiovascular burden and/or in the context of secondary prevention. Furthermore, ethnicity-specific analyses were not performed in the current investigation. Although prior investigations in European-derived, African-derived, and Asian-derived populations have shown consistent effects of PEAR1 rs12041331 genotype with regard to platelet aggregation, the vast majority of ASPREE participants were white (~ 93%). Therefore, caution should be used when extrapolating these results to populations of other ethnic/racial origins, especially given that the minor allele frequency of rs 12041331 varies significantly among such groups. Our study had very few events in the AA homozygote group, which limited the interpretation of results from the additive genetic model used in the primary analysis. To account for this, we conducted analysis using a dominant model, and found similar results. Finally, it is important to note that participants were randomized to low dose aspirin in this study (i.e., 100 mg/daily). If a true interaction between PEAR1 genotype and aspirin use exists, it is possible that such an interaction may be dependent on aspirin dose.
It is also noteworthy that low-dose aspirin did not significantly decrease CVD, MACE, or stroke events vs. placebo in the overall ASPREE trial.21 The lack of an aspirin treatment effect in the ASPREE healthy elderly population must be considered when interpreting the lack of associations found in genetic analysis of PEAR1 rs12041331 in this cohort. It is possible that the selection bias that occurred as a result of the strict ASPREE enrollment criteria, excluding any individuals with a history of diagnosed cardiovascular events, has resulted in ascertainment ofa unique population in which genetic effects are diminished or attenuated. This is indeed consistent with others studies that have shown a diminished effect of genetic risk factors as a function of age in other contexts.35,36
Based on our calculations of minimal detectable differences between rs12041331 genotypes and the end points analyzed in the ASPREE trial (Tables S9, S10), it is possible that larger study populations or meta-analyses may uncover significant effects of PEAR1 and aspirin use on cardiovascular or bleeding events. However, it is also possible that in such larger study populations, investigations may find the effect of rs12041331 moving closer toward zero. Further post hoc analysis of other large aspirin trials is therefore warranted, as new datasets from other large trials become available.
In conclusion, we could not replicate previous findings suggesting that PEAR1 rs12041331 was an important genetic determinant of cardiovascular outcomes. Furthermore, no effects of this variant on clinically significant bleeding was observed in the ASPREE study. Although differences in study designs and participant populations may, in part, explain these divergent results, our data do provide robust evidence that genetic variation in PEAR1 do not contribute to CVD risk in relatively healthy older individuals regardless of aspirin treatment. Additional well-powered investigations in patients with high-risk clinical indications would likely be helpful to determine the role, if any, PEAR1 has on the occurrence of adverse clinical events.
Supplementary Material
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Identification of genetic factors that influence aspirin response may facilitate more personalized antiplatelet treatment, and targeted prevention of cardiovascular disease (CVD). Yet, identification of pharmacogenomic variants of high clinical effect for aspirin has been challenging. Previous studies suggest that genetic variation in the platelet endothelial aggregation receptor 1 (PEAR1) gene results in altered aspirin efficacy. However, association of PEAR1 genotypes with cardiovascular events is less clear.
WHAT QUESTION DID THIS STUDY ADDRESS?
Our study, the largest study of PEAR1 and cardiovascular outcomes to date, examined the effect of rs12041331 genotypes and aspirin use on cardiovascular and bleeding events in a population of 13,547 healthy older individuals without a history of atherothrombotic CVD enrolled in the ASPirin in Reducing Events in the Elderly (ASPREE) trial.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
We observed no association between genotypes and these outcomes, and no interaction with aspirin use in the ASPREE population. Our study indicates PEAR1 genetic variation does not influence aspirin primary prevention in healthy older individuals without a history of cardiovascular events.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Our study represents an important finding that contributes to the evolving pharmacogenomics evidence-base for aspirin, one of the world’s most widely used drugs.
ACKNOWLEDGMENTS
The authors thank the trial staff in Australia and the United States, the participants who volunteered for this trial, and the general practitioners and staff of the medical clinics who cared for the participants.
FUNDING
The ASPREE study was supported by grants from the National Institute on Aging and the National Cancer Institute at the National Institutes of Health (U01AG029824 and R01HL137922), and by grants from the National Health and Medical Research Council of Australia (334047 and 1127060), by Monash University and the Victorian Cancer Agency, and by Bayer AG for the provision of aspirin and placebo. P.L is supported by a National Heart Foundation Future Leader Fellowship and J.P.L is supported by R01HL137922 from the National Institutes of Health.
Footnotes
CONFLICT OF INTEREST
J.P.L. receives grant funding to study the pharmacogenomics of antiplatelet agents. A.R.S. is an employee of Regeneron Pharmaceuticals and receives compensation for his employment. All other authors declared no competing interests for this work.
† These authors should be considered joint first authors
‡ These authors should be considered joint senior authors.
SUPPORTING INFORMATION
Supplementary information accompanies this paper on the Clinical Pharmacology & Therapeutics website (www.cpt-journal.com).
© 2020 The Authors Clinical Pharmacology & Therapeutics © 2020 American Society for Clinical Pharmacology and Therapeutics
References
- 1.Becker DM et al. Sex differences in platelet reactivity and response to low-dose aspirin therapy. JAMA 295, 1420–1427 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Hankey GJ & Eikelboom JW Aspirin resistance. Lancet 367, 606–617 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Larsen SB, Grove EL, Neergaard-Petersen S, Wurtz M, Hvas AM & Kristensen SD Determinants of reduced antiplatelet effect of aspirin in patients with stable coronary artery disease. PLoS One 10, e0126767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faraday N et al. Heritability of platelet responsiveness to aspirin in activation pathways directly and indirectly related to cyclooxygenase-1. Circulation 115, 2490–2496 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Nanda N et al. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. J. Biol. Chem. 280, 24680–24689 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Kauskot A, Di Michele M, Loyen S, Freson K, Verhamme P & Hoylaerts MF A novel mechanism of sustained platelet alphaIIbbeta3 activation via PEAR1. Blood 119, 4056–4065 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Kauskot A et al. PEAR1 attenuates megakaryopoiesis via control of the PI3K/PTEN pathway. Blood 121, 5208–5217 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Johnson AD et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat. Genet. 42, 608–613 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qayyum R et al. Genome-wide association study of platelet aggregation in African Americans. BMC Genet. 16, 58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrera-Galeano JE et al. A novel variant in the platelet endothelial aggregation receptor-1 gene is associated with increased platelet aggregability. Arterioscler. Thromb. Vasc. Biol. 28, 1484–1490 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faraday N et al. Identification of a specific intronic PEAR1 gene variant associated with greater platelet aggregability and protein expression. Blood 118, 3367–3375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keramati AR et al. Targeted deep sequencing of the PEAR1 locus for platelet aggregation in European and African American families. Platelets 30, 380–386 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis JP et al. Pharmacogenomic polygenic response score predicts ischemic events and cardiovascular mortality in clopidogrel-treated patients. Eur. Heart J. Cardiovasc. Pharmacother. 10.1093/ehjcvp/pvz045. [e-pub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M et al. Association of PEAR1 rs12041331 polymorphism and pharmacodynamics of ticagrelor in healthy Chinese volunteers. Xenobiotica 47, 1130–1138 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Xiang Q, Cui Y, Zhao X & Zhao N Identification of PEAR1 SNPs and their influences on the variation in prasugrel pharmacodynamics. Pharmacogenomics 14, 1179–1189 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Izzi B et al. Allele-specific DNA methylation reinforces PEAR1 enhancer activity. Blood 128, 1003–1012 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Izzi B et al. Variation of PEAR1 DNA methylation influences platelet and leukocyte function. Clin. Epigenetics 11, 151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNeil JJ et al. Baseline characteristics of participants in the ASPREE (ASPirin in reducing events in the elderly) study. J. Gerontol. A Biol. Sci. Med. Sci. 72, 1586–1593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNeil JJ et al. Effect of aspirin on all-cause mortality in the healthy elderly. N. Engl. J. Med. 379, 1519–1528 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNeil JJ et al. Effect of aspirin on disability-free survival in the healthy elderly. N. Engl. J. Med. 379, 1499–1508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNeil JJ et al. Effect of aspirin on cardiovascular events and bleeding in the healthy elderly. N. Engl. J. Med. 379, 1509–1518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das S et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandenbriele C et al. Dextran sulfate triggers platelet aggregation via direct activation of PEAR1. Platelets 27, 365–372 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Xiang Q, Zhou S, Lewis JP, Shuldiner AR, Ren G & Cui Y Genetic variants of PEAR1 are associated with platelet function and antiplatelet drug efficacy: a systematic review and meta-analysis. Curr. Pharm. Des. 23, 6815–6827 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Zhan Q, Ma X & He Z PEAR1 suppresses the proliferation of pulmonary microvascular endothelial cells via PI3K/AKT pathway in ALI model. Microvasc. Res. 128, 103941 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Vandenbriele C et al. Platelet endothelial aggregation receptor-1: a novel modifier of neoangiogenesis. Cardiovasc. Res. 108, 124–138 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Wu HH et al. Glial precursors clear sensory neuron corpses during development via Jedi-1, an engulfment receptor. Nat. Neurosci. 12, 1534–1541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan CS et al. The adaptor protein GULP promotes Jedi-1-mediated phagocytosis through a clathrin-dependent mechanism. Mol. Biol. Cell 25, 1925–1936 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis JP et al. Genetic variation in PEAR1 is associated with platelet aggregation and cardiovascular outcomes. Circ. Cardiovasc. Genet. 6, 184–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang WY et al. PEAR1 is not a major susceptibility gene for cardiovascular disease in a Flemish population. BMC Med. Genet. 18, 45 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nie XY et al. Genetic mutations in PEAR1 associated with cardiovascular outcomes in Chinese patients with acute coronary syndrome. Thromb. Res. 163, 77–82 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Xu K et al. Impact of platelet endothelial aggregation receptor-1 genotypes on platelet reactivity and early cardiovascular outcomes in patients undergoing percutaneous coronary intervention and treated with aspirin and clopidogrel. Circ. Cardiovasc. Interv. 12, e007019 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Sokol J, Skerenova M, Ivankova J, Simurda T & Stasko J Association of genetic variability in selected genes in patients with deep vein thrombosis and platelet hyperaggregability. Clin. Appl. Thromb. Hemost. 24, 1027–1032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pi L et al. A PEAR1 polymorphism (rs12041331) is associated with risk of coronary artery aneurysm in Kawasaki disease. Ann. Hum. Genet. 83, 54–62 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Kuchenbaecker KB et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317, 2402–2416 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Moller P et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: a report from the Prospective Lynch Syndrome Database. Gut 67, 1306–1316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


