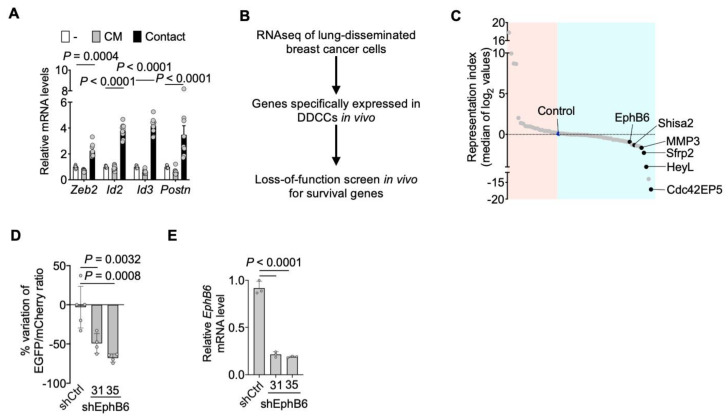
Figure 1.
EphB6 supports the survival of disseminated dormant breast cancer cells. (A) RT-qPCR analysis of representative genes of the dormancy response. D2.0R-EGFP cells have been cultured alone, cocultured with AT1-like cells (Contact), or with conditioned medium from AT1-like cells (CM). Mean normalized pooled samples from n = 3 independent experiments. 2 way ANOVA, multiple comparisons. Error bars: SD. (B) Workflow of the loss-of-function screen in vivo for survival genes in disseminated dormant cancer cells (DDCCs) [8]. (C) Representation scores for each gene included in the screen, calculated from the fold change of representation of each shRNA relative to pre-injection abundance. On the light blue side, there are genes whose downregulation leads to a reduced representation of the clones. Black dots indicate genes (ranking among the top 10 genes) with a consistent effect of at least 2 out of 3 shRNAs included in the screening. (D) D2.0R-EGFP cells stably expressing the indicated shRNA were injected intravenously together with an equal amount of D2.0R-mCherry-shCtrl cells as an internal control. After 3 weeks the amount of surviving D2.0R cells was measured and the ratio EGFP/mCherry calculated. n = 5 mice for shCtrl cells, n = 4 mice for each shEphB6 sequence. One-way ANOVA test. Mean with SD. (E) qPCR analysis of Ephb6 mRNA in D2.0R-EGFP cells stably expressing shRNA. One-way ANOVA test. Mean with SD.