Significance
We identify the replicative polymerase polymerase δ (Pol δ) as a promoter of aberrant DNA double-strand break end-joining events in human cells, including chromosomal translocations between targeted double-strand breaks. These studies identify a previously unknown role for Pol δ in human cells of multiple lineages and suggest a mechanism of Pol δ-dependent mutagenesis.
Keywords: polymerase δ, POLD2, POLD1, translocation, nonhomologous end-joining
Abstract
Recent studies have implicated DNA polymerases θ (Pol θ) and β (Pol β) as mediators of alternative nonhomologous end-joining (Alt-NHEJ) events, including chromosomal translocations. Here we identify subunits of the replicative DNA polymerase δ (Pol δ) as promoters of Alt-NHEJ that results in more extensive intrachromosomal mutations at a single double-strand break (DSB) and more frequent translocations between two DSBs. Depletion of the Pol δ accessory subunit POLD2 destabilizes the complex, resulting in degradation of both POLD1 and POLD3 in human cells. POLD2 depletion markedly reduces the frequency of translocations with sequence modifications but does not affect the frequency of translocations with exact joins. Using separation-of-function mutants, we show that both the DNA synthesis and exonuclease activities of the POLD1 subunit contribute to translocations. As described in yeast and unlike Pol θ, Pol δ also promotes homology-directed repair. Codepletion of POLD2 with 53BP1 nearly eliminates translocations. POLD1 and POLD2 each colocalize with phosphorylated H2AX at ionizing radiation-induced DSBs but not with 53BP1. Codepletion of POLD2 with either ligase 3 (LIG3) or ligase 4 (LIG4) does not further reduce translocation frequency compared to POLD2 depletion alone. Together, these data support a model in which Pol δ promotes Alt-NHEJ in human cells at DSBs, including translocations.
Translocations are genetic rearrangements involving the fusion of heterologous chromosomes (1) and can be initiated by two or more DNA double-strand breaks (DSBs) (2, 3). DSBs in human cells are repaired by multiple pathways with distinct genetic requirements. The first pathway, homology-directed repair (HDR), is active during the S/G2 phase of the cell cycle, when sister chromatids are present to template DNA synthesis. In the first step of HDR, the DSB ends undergo 5′-to-3′ single-strand resection that involves the Mre11/Rad50/Nbs1 (MRN) complex and the endonuclease CtIP (4). The RPA complex binds the exposed single-stranded DNA and is then exchanged for RAD51 by BRCA2 (5). The RAD51 nucleoprotein filament then facilitates strand invasion into a homologous duplex that serves as a repair template. Multiple DNA polymerases, including the replicative polymerases (Pol δ and Pol ε) and translesion DNA polymerases can participate in DNA synthesis during HDR in mammalian cells (6, 7), which is followed by ligation of the ends.
The second pathway, classic nonhomologous end-joining (C-NHEJ), is active throughout the cell cycle and therefore responsible for the repair of most DSBs in somatic cells (5). In C-NHEJ, the DSB is bound by the Ku70/Ku80 (Ku) heterodimer, resulting in recruitment of DNA-dependent protein kinase and the end-bridging factors XLF and PAXX (8). End-processing factors, including Artemis and DNA polymerases μ and λ, can also be recruited for end-resection and gap-filling (9, 10). The XRCC4/LIGIV complex is recruited and ligates both strands (11).
The third type of repair, alternative NHEJ (Alt-NHEJ), is often described as a back-up end-joining process, as it resolves a greater fraction of DSBs when C-NHEJ is compromised (12). Alt-NHEJ is also more error-prone than C-NHEJ and appears to contribute to mutagenesis in many types of cancer cells (13, 14). A large number of potentially redundant factors may participate in Alt-NHEJ, including CtIP, MRN, Pol θ (encoded by POLQ), poly(ADP-ribose) polymerase 1 (PARP1), and Ligases 1 and 3 (Lig1/3) (4, 12, 15–21). The large number of factors suggests that the “pathway” is actually a combination of potential mediators that compete at the break. As a result, context-specific, lineage-driven, and stochastic effects may each influence which factors ultimately mediate Alt-NHEJ at a given DSB.
Similar to HDR, the first step of Alt-NHEJ can involve resection of the DNA ends to single-strands by MRN and CtIP. During HDR, single-strand resection typically extends for kilobases but the extent of resection is limited in G0/G1 phases of the cell cycle by 53BP1. Thus, Alt-NHEJ occurring during G0/G1 involves short 3′ overhangs that may anneal at sites of microhomology (4). Previous studies with mammalian cells have described varying lengths of microhomology characteristic of Alt-NHEJ junctions [e.g., ≥2 bp (22), ≥3 bp (23), ≥5 bp (24), and 2 to 6 bp (25)], suggesting that there is no absolute requirement for a given length across contexts.
After annealing, DNA polymerases and ligases are needed to fill the gaps before end ligation. The role of polymerases in Alt-NHEJ (and translocation formation) remains poorly understood. Pol θ is thought to play a role in Alt-NHEJ by facilitating DNA synthesis from annealed microhomologies, and loss of Pol θ led to reduced frequency of Cas9-induced translocations (17) and increased frequency of spontaneous IgH/Myc translocations in mouse cells (26). A recent study showed that loss of Pol β can also reduce translocation frequency between endonuclease-induced breaks in human cells (27).
Translocation junctions in mammalian cells tend to have longer deletions and increased use of microhomology compared to repair at single DSBs, suggesting that Alt-NHEJ is involved. Initial studies on the mechanisms of translocation formation were performed in mouse embryonic stem cells (mESCs) containing a translocation reporter that reconstitutes a neomycin resistance gene after cleavage of chromosomes 14 and 17 by the I-SceI meganuclease (22, 28, 29). mESCs lacking either Ku or Xrcc4/Lig4 had increased translocation frequencies, suggesting that these factors suppress translocations rather than promoting them (28). Furthermore, in the absence of C-NHEJ factors, translocation junctions contained longer deletions and an increased usage of microhomology in the final repair products, consistent with Alt-NHEJ mediating translocations in these cells. Additional studies in mESCs demonstrated that loss of CtIP, Parp1, Lig3, and Lig1 can each result in reduced translocation frequency, further implicating these factors in the Alt-NHEJ that mediates translocation formation (22, 29).
A subsequent study in human cells painted a more complex picture. Human cell lines depleted of LIG4 had reduced translocation frequency and depletion of LIG3 only decreased translocations in LIG4-depleted cells (23). The mechanisms and cell type-specificity behind these differential end-joining requirements for translocation formation in mouse and human cells remain poorly defined (23).
We previously reported a screen of short hairpin RNA (shRNA) against 169 DNA repair-related genes in human cells to identify factors that modulate translocation frequency (30). We used stringent criteria to define “hits” and validated each in multiple human cell lines. Knockdown of the SUMO E2 enzyme UBC9 or RAD50 increased translocation frequency, so we categorized these factors as translocation suppressors. Conversely, knockdown of 53BP1, DNA damage-binding protein 1 (DDB1), or PARP3 decreased translocation frequency, so we categorized these as translocation promoters (30).
We noted that POLD2, an accessory subunit of the replicative polymerase Pol δ, nearly scored in the screen as a promoter of chromosomal translocations. In budding yeast, Pol δ promotes both Alt-NHEJ and microhomology-mediated chromosomal translocations (31) but this has not been assessed in mammalian cells. Here, we demonstrate that Pol δ plays a role during Alt-NHEJ in human cells and that Pol δ subunits promote translocations. Based on these findings, we propose a model in which Pol δ exonuclease and polymerase activity promote Alt-NHEJ after annealing of sequences with microhomology.
Methods
Analysis of shRNA Screen.
We previously screened (30) a 169 factor DNA repair library in which each gene was represented by four to five shRNA. In that study, we identified five genes that reached our threshold of ≥two shRNA for a given gene exhibiting ≥twofold change in the same direction compared to controls. We defined this threshold based on mean values across all seven replicates. For additional stringency, we required that hits exhibit significant hairpin enrichment in ≥four of seven individual screening replicates. In the present report, we maintained these criteria to identify genes that have ≥two shRNA exhibiting ≥twofold change in the same direction compared to controls based on a mean from all seven replicates. However, we relaxed only the threshold that the gene should additionally exhibit significant enrichment in ≥three of seven individual screening replicates.
Cell Culture.
HeLa, 293T, and U2OS cells were all cultured in DMEM supplemented with penicillin/streptomycin, 200 mM glutamine, and 10% fetal bovine serum (Sigma). All cells were cultured under normal oxygen conditions (21% O2, 5% CO2, 37 °C). For cells exposed to hydroxyurea, cells were treated with either 5 mM hydroxyurea or DMSO for 24 h before preparation of cell lysate.
Lentiviral Transductions.
Cells were plated at 60,000 cells per well of 12-well tissue culture plates and 5 μL of lentivirus and polybrene to a final concentration of 10 μg/mL were added. Cells were spun at 1,178 × g for 15 min and returned to incubator overnight. Twenty-four hours after transduction, cells were cultured in 1 μg/mL puromycin for 48 h.
Constructs.
The FLAG-HA-POLD2 plasmid was constructed using pDONR221 HsCD00044072 (https://plasmid.med.harvard.edu/PLASMID/Home.xhtml) as a backbone. Mutations causing small-interfering RNA (siRNA) resistance were incorporated into the ORF using restriction enzyme cloning (NheI and AgeI) and a custom gBlock (Integrated DNA Technologies, IDT). The FLAG-HA tag was incorporated using PCR and the resulting construct pDONR221-FLAG-HA-POLD2mut was moved into pCW107 using Gateway cloning (Invitrogen) for lentiviral production.
The POLD1 human ORF was obtained from the CCSB Human ORFeome Collection (Dana-Farber Cancer Institute) and subcloned into pCMV using Gateway Cloning (Invitrogen). Using a custom gBlock (IDT), silent mutations leading to siRNA resistance (to siRNA J-019687-06, J-019687-07) were incorporated into the ORF using restriction enzyme cloning (BamHI and BstZ17I). The D316G exonuclease-deficient mutation and the del605S polymerase dead mutations were also incorporated using custom gBlocks (IDT). Finally, the resulting constructs POLD1 WT, POLD1 D316, and POLD1 del605S were cloned into pCW107 using Gateway Cloning (Invitrogen). All constructs were verified by Sanger Sequencing (Eton Bioscience). The 53BP1−/− and LIG4−/− 293T cells were described in Day et al. (30).
siRNA, cDNA Transfection, and Nucleofection.
Initial siRNA transfections were performed with Dharmafect (Dharmacon) according to the manufacturer’s instructions. For this, 5 × 104 cells were plated in 12-well plates and each well was transfected with 2 µL of Dharmafect reagent and contained a final siRNA concentration of 31.25 nM in a total volume of 1 mL siRNA used are as follows: ON-TARGETplus Human POLD2 #1 (J-020131-09) siRNA, ON-TARGETplus Human POLD2 #2 siRNA (J-020131-10), ON-TARGETplus Human POLQ Pool (L-015180-01) siRNA, ON-TARGETplus Human POLD1 #1 siRNA (J-019687-06), ON-TARGETplus Human POLD1 #2 siRNA (J-019687-07), ON-TARGETplus Human POLB siRNA (J-005164-06), ON-TARGETplus Human POLQ Pool siRNA (L-015180-01), ON-TARGETplus Human LIG3 siRNA (J-009227-06), and ON-TARGETplus Nontargeting (D-001810-02) siRNA.
For CRITR (CRISPR/Cas9-based translocation reporter) assays, the cells were transfected a second time with the indicated siRNAs and 5 µg of a plasmid encoding the CD4 and CD71 gRNAs. Forty-eight hours following the second transfection, the cells were harvested using Cellstripper (Cat# 25-056CI, Corning Life Sciences) and washed with FACS buffer (PBS pH 7.4, 2 mM EDTA, 1% FBS). Cells were then Fc Blocked using Human TruStain FcX (Cat# 422302 Biolegend) according to the manufacturer’s instructions. Cells were washed with FACS buffer and then stained with CD4-PE (Clone M-T466, Miltenyi) and incubated on ice for 10 min. Cells were washed with FACS buffer and analyzed by flow cytometry. Guide RNAs (gRNA) can be found in SI Appendix, Table S5. For U2OS-DRGFP cells, siRNAs and pCAGGS or pCAGGS-ISceI (pCBaSce) were introduced by Nucleofection Kit V (Lonza) according to the manufacturer’s instructions. Forty-eight hours following the nucleofection, cells were harvested for flow cytometry. To monitor gene targeting, 293T cells containing a lentivirally integrated GFP were transfected with siRNAs and 72 h later transfected with siRNA, Cas9/gRNA expression vector, and a linear repair template. After 10 d, the cells were analyzed by flow cytometry. The repair template was generated by PCR amplification of a gBlock (IDT) with the sequence: AGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGGAGAGGGCGATGCCACCTACGGCAAACTGACCCTGAAGTTCATCTGCACCACTGGCAAGCTGCCCGTGCCCTGGCCAACCCTCGTGACCACCCTGAGCCACGGGGTGCAGTGCTTCAGCCGCTACCCAGACCACATGAAGCAGCACGACTTTTTCAAGTCCGCCATGCCCGAAGGCTATGTCCAGGAGCGCACCATCTTCTTTAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTTCGAGGGCG and primers: Forward GCGACGTAAACGGCCACAAG and Reverse GCCCTCGAACTTCACCTCG.
For POLD1 complementation experiments, 293T cells were transfected with pCW107-POLD1 WT, pCW107-POLD1 D316G, or pCW107-POLD1 del605S constructs using Lipofectamine 3000 (ThermoFisher Scientific) according to the manufacturer’s instructions. Transduced cells were selected 48 h following the transfection with 1 μg/mL puromycin. Data generated were the results of three independent experiments.
ZiTR and CRITR Assays.
Zinc finger nuclease (ZFN) translocation reporter (ZiTR) and CRITR assays were performed as previously described (30). Data generated were the results of three independent experiments. gRNA sequences for CRITR can be found in SI Appendix, Table S5.
Proximity Ligation Assays.
For proximity ligation assays (PLAs), 50,000 U2OS cells were seeded on coverslips and allowed to adhere overnight. The next day cells were treated with 10 Gy of ionizing radiation (IR) or mock-treated and allowed to recover for 2 h before fixation in 4% paraformaldehyde for 15 min at room temperature. Cells were then stored in 70% ethanol at −20 °C overnight. The following morning, cells were permeabilized for 10 min at room temperature in 0.1% Triton X-100 in PBS. The samples were then processed using the Red DuoLink Proximity Ligation Assay Kit (Sigma, DUO92101) according to the manufacturer’s instructions using anti-POLD2 (HPA026745, Sigma and sc-390583, Santa Cruz Biotechnology), anti–γ-H2AX (JBW301, Millipore and 07-164, Millipore), and anti-53BP1 (NB100-904, Novus Biologicals), anti-POLD1 (sc-17777, Santa Cruz Biotechnology), anti-POLβ (ab26343, Abcam), and anti-POLθ (ab80906, Abcam). All antibodies were used at 1:500 dilution. Images were acquired on an inverted Leica DMI6000 Laser Scanning Confocal Microscope using a 40× Plan Apo oil-immersion objective using lasers at 405 nm and 594 nm. All confocal microscopy images for this study were acquired in the Confocal and Light Microscopy Core facility at the Dana-Farber Cancer Institute. At least 90 cells were counted for each condition in each biological replicate. Data generated were the results of two independent experiments.
Chromatin Immunoprecipitation.
Chromatin samples were prepared and immunoprecipitated using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling) by the manufacturer’s instructions. Briefly, stable POLD2-HA-FLAG expressing 293T cells were transfected with p84-ZFN (10 μg) using Lipofectamine 2000 (Life Technologies) or Lipofectamine only (no cut control) and chromatin was isolated 18 h posttransfection. The chromatin was immunoprecipitated overnight at 4 °C with 7 μg anti-HA (Abcam, 9110). POLD2-HA recruitment was determined by qPCR using primer sets proximal to the p84-ZFN cut site at the AAVS1 locus (as listed in ref. 32). Data generated were the results of four independent experiments.
Next-Generation Sequencing.
Genomic DNA (gDNA) from CRITR assays was isolated using the Qiagen DNA/Blood Mini kit. Eight 100-ng gDNA aliquots were used for amplification of CD4 and ESR1 products with Accupower PCR PreMix (Bioneer). Cycling conditions were 95 °C for 3 min, and then 27 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s. After amplification, identical reactions were pooled and purified using the Qiagen PCR purification kit. Purified products were visualized on an agarose gel to ensure the presence of a single band. Next-generation sequencing was performed using a MiSeq System (Illumina) by the Center for Computational and Integrative Biology DNA Core Facility at Massachusetts General Hospital (Cambridge, MA) using a MiSeq v2 chemistry 300-cycle kit. Data generated were the results of three independent experiments.
For RNA-sequencing experiments, HeLa cells were transiently transfected with either nontargeting control siRNA, siPOLD2 #1 (Dharmacon, J-020131-09) or siPOLD2 #2 (J-020131-10). The cells were transiently transfected again with same siRNA 72 h after the first transfection. Total RNA was harvested 48 h after the second transfection and total RNA was prepared using the RNeasy Plus Mini Kit (Qiagen, 74104). RNA was subjected to Illumina 2 × 150 bp RNA sequencing at the Koch Institute at MIT (Cambridge, MA). Raw FASTQ files were mapped using Bowtie and normalized using the Bioconductor package DESeq2 in R (33). Differential expression analysis was performed using DESeq2.
Bioinformatics.
Raw reads from paired end sequencing were trimmed to amplicon primer sequences and merged into single reads using Geneious v10.1.3. Junctions lacking 20 bp of reference sequence immediately internal to primer sequences were discarded as PCR artifacts. Junctions were trimmed to common starting and ending coordinates, mapped to the CD4 or ESR1 reference sequence, and exported as SAM files using Geneious v10.1.3. Using Hi-FiBR (34), CIGAR strings were analyzed to classify sequences as exact, deletion, complex, or insertion, and to determine the extent of deletions, insertions, and microhomology usage within each sequence.
For the analysis of repair classes and lengths of deletions, insertions, and microhomology, junctions with ≥10 reads were analyzed to minimize the effects of PCR artifacts observed in the uncut control. We excluded exact reads from the analysis as we cannot distinguish between perfect repair and uncut. The normalized percentage of each repair class was determined by dividing the number of reads corresponding to deletion, complex, or insertion by the total reads analyzed. Deletion, insertion, and microhomology lengths were calculated by multiplying the read counts associated with each deletion, insertion, or microhomology length, and dividing by the total read count associated with deletion and complex classes, insertion and complex classes, or deletion class, respectively.
For analysis of templated insertions, all insertions >20 bp were considered for further analysis. To control for PCR artifacts seen in the uncut control, sequences containing a deletion of n bp on the left or right side of the break and an insertion of n or n−1 were excluded from the templated insertion analysis. Additionally, any junctions called with deletions that would eliminate a primer sequence were removed from the dataset. The remaining sequences were submitted to BLAST under default or short sequence parameters against a local Homo sapiens refseq genomic GRCh38.p12 database ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/vertebrate_mammalian/Homo_sapiens/latest_assembly_versions/ and the top hits were exported as XML files for further analysis.
Limiting Dilution PCR and Translocation Junction Analysis.
gDNA was isolated from 293T cells at the end of the CRITR assay using the QiaAMP DNA Blood Mini Kit (Qiagen). gDNA template was diluted to 1.5 ng per reaction to achieve fewer than one distinct PCR products for every three reactions. A nested PCR strategy was used to isolate individual junctions. Both reactions used the AccuPower PCR PreMix (Bioneer) and the cycling conditions were 95 °C for 1 min, followed 2 cycles of 95 °C for 30 s, 74 °C for 30 s, 72 °C for 30 s; 2 cycles of 95 °C for 30 s, 70 °C for 30 s, 72 °C for 30 s; 2 cycles of 95 °C for 30 s, 66 °C for 30 s, 72 °C for 30 s; 2 cycles of 95 °C for 30 s, 62 °C for 30 s, 72 °C for 30 s; 2 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s; and 25 cycles of 95 °C for 30 s, 70 °C for 30 s, 72 °C for 30 s. A 1.0-μL aliquot of the first reaction was used as the template for the second reaction. A small aliquot of each second reaction was analyzed by agarose gel to determine if it had a translocation product and those reactions with distinct PCR products were treated with EXOSap-IT (Affymetrix) and analyzed by Sanger sequencing. Data generated were the results of three independent experiments. The nested PCR primers were Outer F: AGGTTGGGAGGGTGGGTAAG, Outer R: AGGGGATGAAAGAGGGGAAG, Inner F: CGGCTCTTTTCGGATAACGC, Inner R: GCATTCTTGACCCTTCCCCA. The sequencing primer was TCGGATAACGCTTTCCCTCTGC.
Cell Cycle Analysis.
Forty-eight hours following the second siRNA transfection, cells were processed using the Click-iT Plus EdU Pacific Blue Flow Cytometry Assay Kit (ThermoFisher Scientific C10636) according to the manufacturer’s instructions. Data generated were the results of three independent experiments.
Cell Proliferation Assay.
Twenty-five thousand 293T cells were seeded into a 24-well plate and transfected in triplicate with the appropriate siRNA using Dharmafect (Horizon Discovery). The cells were harvested using Cellstripper (Corning) 2, 3, and 5 d following siRNA transfection. Then 100 μL of each of the single-cell suspensions were transferred in duplicate to a 96-well plate and a fixed-volume of cells was counted by flow cytometry using a FACSCanto II flow cytometer (BD Biosciences).
Western Blotting.
Cells were washed with ice-cold PBS and lysed on ice for 30 min with RIPA lysis buffer supplemented with protease inhibitors (Complete Mini, Roche). Protein concentration was determined by the Bradford Protein Assay Kit (Bio-Rad) and a calibration standard curve created from BSA. The samples were prepared for loading by adding 4X Sample Buffer (Life Technologies) and heating the samples at 90 °C for 10 min. Total proteins were separated by SDS–PAGE on 4 to 12% Bis-Tris gradient gels (Life Technologies). Proteins in the gel were electrophoretically transferred to nitrocellulose membrane (Bio-Rad), Ponceau-stained, and photographed. The membrane was then blocked in 0.5% milk with Tris-Buffered Saline Triton X-100 buffer (100 mM Tris, pH 7.4, 500 mM NaCl, 0.1% Triton X-100) (TBST). Membranes were incubated with primary antibodies in 0.5% milk in TBST overnight at 4 °C. HRP-conjugated secondary antibody incubation was performed for 2 h at room temperature in 0.5% milk in TBST and signals were visualized by ECL (Perkin-Elmer). Primary antibodies used in this study were as follows: POLD2 (HPA026745 Sigma), POLD1 (ab10862 Abcam), POLD1 (sc-17777 Santa Cruz Biotechnology), POLD3 (A301-244A-M Bethyl Laboratories), p-RPA (A300-245A Bethyl Laboratories), RPA32 (A300-244A Bethyl Laboratories), p-ATR (sc109912 Santa-Cruz), ATR (A300-137A Bethyl Laboratories), anti–α-Tubulin (T9026 Sigma Aldrich), anti-GAPDH (5174 Cell Signaling Technology), anti–Myc-tag (2276 Cell Signaling Technology), and anti-LIG4 (SC-271299 Santa Cruz Biotechnology). All antibodies were used at 1:1,000 dilutions.
Real-Time qPCR.
Total DNA was extracted from 293T cells using the RNeasy Mini Kit (Qiagen) 24 h following transfection of CRISPR/Cas9 constructs to measure Cas9 cutting efficiency. For knockdown validations, total RNA was extracted 48 h following the second siRNA transfection. Single-stranded cDNA was synthesized using 500 ng of total RNA with the iScript cDNA Synthesis Kit (Bio‐Rad) according to the manufacturer’s instructions. Real-time qPCR (RT-qPCR) was performed on a CFX96 Real Time System (Bio-Rad) machine using iTaq UniverSYBR Green (Bio-Rad). RT-qPCR conditions were: 50 °C for 10 min; 95 °C for 10 min; 39 cycles of 95 °C for 10 s and 60 °C for 30 s. Data generated were the results of two independent experiments. The RT-qPCR primers are listed in SI Appendix, Table S5. Data analysis was performed using the ΔΔCT-method. The housekeeping gene GAPDH was used to normalize the data.
Statistics.
Translocation frequencies, direct repeat (DR)-GFP, gene targeting assays, qPCR, and PLA results were analyzed using a one-way ANOVA with Tukey’s correction. Hi-FiBR results were analyzed using a one- or two-way ANOVA with Tukey’s correction. Differential expression of bulk RNA sequencing (RNA-seq) between experimental conditions was determined using raw count data and normalization procedures within the DESeq2 package in R based on a negative binomial distribution. The false-discovery rate by the Benjamini and Hochberg method was used to adjust for multiple comparisons.
Results
POLD2 Promotes Translocations in Multiple Human Cell Types.
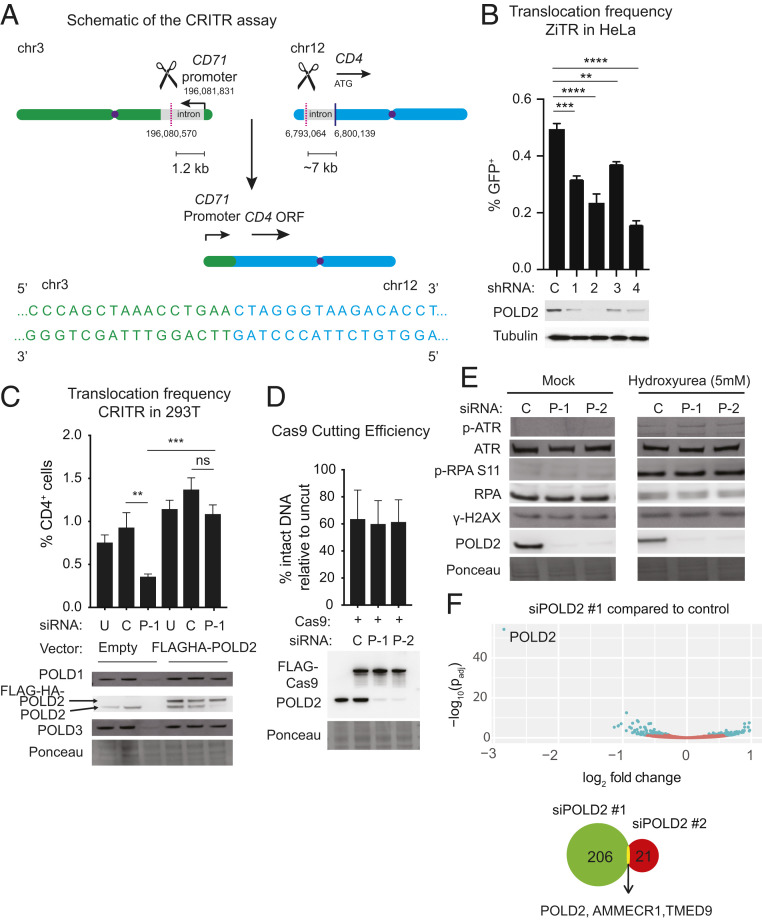
We reanalyzed data from our previous screen (30) for factors affecting translocation frequency using a ZiTR in human A549 lung adenocarcinoma cells. Using less stringent criteria, POLD2, a regulatory subunit of the replicative polymerase Pol δ, scored as a promoter of translocations (Methods). To confirm the effects, we assayed translocation frequency after POLD2 knockdown in HeLa cervical carcinoma cells using the ZiTR (SI Appendix, Fig. S1A) and in 293T embryonic kidney cells using a CRITR that results in aberrant expression of CD4 by juxtaposing the CD4 coding sequence downstream of the CD71 promoter (Fig. 1A and SI Appendix, Fig. S1B) (30). POLD2 depletion with siRNA reduced translocations 50 to 70% using both ZiTR and CRITR (Fig. 1 B and C). Expression of an siRNA-resistant POLD2 cDNA restored translocation frequency to basal levels (Fig. 1C), consistent with the siRNA effect on translocations directly involving depletion of POLD2.
Fig. 1.
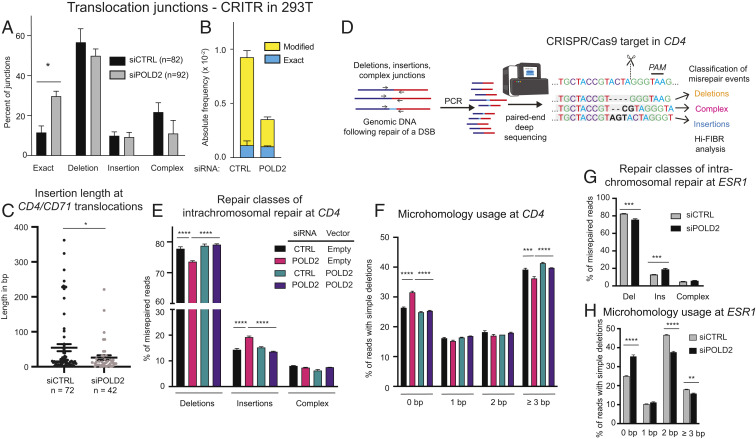
POLD2 promotes chromosomal translocations. (A) Schematic of the CRITR assay (30). (Upper) Endogenous CD71 and CD4 loci showing the orientation of the CD71 promoter and the CD4 ORF on human chromosomes 3 (chr3, green) and 12 (chr12, blue), respectively. Scissors and pink dotted line indicate CRISPR/Cas9 cut site. (Lower) Sequence of translocation junction without any end processing (referred to as “exact”). Translocation juxtaposes the CD71 promoter with the CD4 ORF leading to the expression of the CD4 gene, which is quantified by flow cytometry. Chromosome coordinates correspond to Human Dec. 2013 (GRCh38/hg38, University of California, Santa Cruz [UCSC] Genome Browser). (B) Validation of POLD2 as a genetic factor influencing chromosomal translocations by measuring translocation frequency 48 h after AAVS1 ZFN adenovirus infection into HeLa ZiTR cells (30) stably transduced with one of four different shRNA against POLD2 (1–4) or control (C). (C) Translocation frequency in CRITR assay using 293T cells stably transduced with siRNA-resistant FLAG-HA-POLD2 or empty vector. Cells were transfected with indicated siRNA and 72 h later transfected with the same siRNA and Cas9/gRNA. Cells were collected after 48 h for flow cytometry and immunoblotting. (D) Cas9 cutting efficiency at the CD4 locus. Cells were transfected with siRNA targeting POLD2 or control as in C and then harvested 24 h after Cas9/gRNA transfection for qPCR with primers across the CD4 cleavage site. (E) Immunoblotting in HeLa cells after exposure to vehicle or hydroxyurea (HU) for 24 h. In A–D, U, Untransfected; C, Control; P-1, siPOLD2 #1; P-2, siPOLD2 #2. Data are presented as mean ± SE (SE) of n = 3. P values calculated using a one-way ANOVA with Tukey’s correction. **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant. (F) Differentially expressed genes following siPOLD2 #1. Genes with differential expression (Padj < 0.05) are shown in blue; 206 genes were differentially expressed (P < 0.05) with siPOLD2 #1 relative to siCont and 21 genes were differentially expressed with siPOLD2 #2 relative to siCont. Venn diagram indicates overlap in genes differentially expressed across the two different POLD2 siRNA.
One issue that has not been addressed in several studies is whether loss (by RNAi-mediated depletion or genetic disruption) of candidate factors may be affecting translocation frequency through secondary effects. These could include effects on endonuclease-mediated cleavage, cell cycle progression, gene transcription, DNA replication, or DNA damage at additional sites in the genome. For example, knockdown of essential factors like CtIP is likely to affect cell cycle progression and DNA replication (35). Importantly, POLD2 depletion did not affect Cas9 expression or Cas9 cutting efficiency (Fig. 1D). POLD2 depletion also induced little or no change in the cell cycle distribution (SI Appendix, Fig. S1C) or in the replication stress markers phospho-RPA and phospho-ATR and the DSB marker γH2AX, either in the presence or absence of the replication stressor hydroxyurea (5 mM) (Fig. 1E). These results are consistent with studies in transformed cells, such as HeLa and U2OS osteosarcoma cells, showing that depletion of the Pol δ subunit POLD3 elicits only mild changes in replication stress and spontaneous DSBs without causing a cell cycle delay (36, 37). They are also consistent with studies in yeast showing that strains with low Pol δ can achieve rapid growth (38).
To further determine whether POLD2 knockdown has nonspecific effects on cell state, we performed RNA-seq of POLD2-depleted HeLa cells using two separate POLD2-directed siRNA and controls. Strikingly, only three genes (POLD2, AMMECR1, TMED9) were significantly differentially expressed (Padj < 0.05) by both siRNA (Fig. 1F). Neither AMMECR1 nor TMED9 has previously been implicated in DNA end-joining. Together, these data strongly support the conclusion that POLD2 directly promotes translocations.
POL δ Polymerase and Exonuclease Activities Promote Translocations.
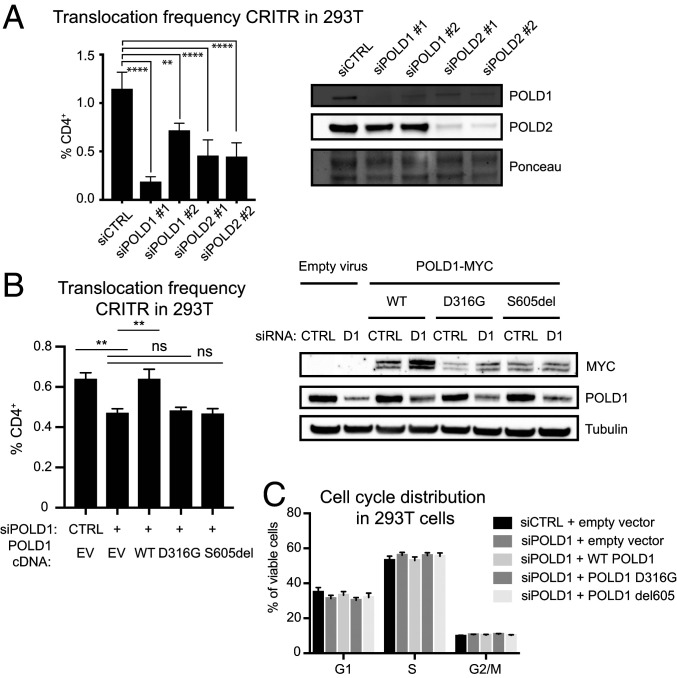
POLD2 depletion led to nearly complete loss of the Pol δ subunits POLD1 and POLD3, which was rescued by expressing siRNA-resistant POLD2 (Fig. 1C). Therefore, we hypothesized that the effects on translocation formation observed with POLD2 knockdown involve the Pol δ holoenzyme. Indeed, depletion of the catalytic subunit POLD1 reduced translocation frequency to a similar extent as POLD2 knockdown (Fig. 2A).
Fig. 2.
Pol δ function promotes chromosomal translocations. (A) Translocation frequency in the CRITR assay (30) using 293T cells after depletion of POLD1 or POLD2. siCTRL indicates nontargeting control. Cells were transfected with the indicated siRNA at 0 h and at 72 h they were transfected with the same siRNA and Cas9/gRNA. Cells were collected after 48 h for flow cytometry and immunoblotting. (B) Translocation frequency in the CRITR assay using 293T cells stably transduced with empty vector (EV) or Myc-tagged, siRNA-resistant POLD1 WT, D316G, or S605del. Cells were transfected with siCTRL or siPOLD1 #2 (D1) and 72 h later transfected with the same siRNA and Cas9/gRNA. Cells were collected after 48 h for flow cytometry and immunoblotting. (C) Cell cycle dynamics for 293T cells transfected with siCTRL or siPOLD1 and transduced with siRNA-resistant POLD1 constructs, as indicated. Data are presented as mean ± SE of n = 3 independent experiments. P values calculated using a one-way ANOVA with Tukey’s correction. No statistically significant differences were observed in C. **P < 0.01, ****P < 0.0001.
POLD1 has both DNA synthesis and exonuclease activities that could contribute to its role in translocation formation. An in-frame deletion of serine 605 (S605del) abolishes POLD1 DNA synthesis activity (39) and a D316G substitution in the exonuclease domain (40) eliminates exonuclease and proofreading activity. We performed knockdown of POLD1 and replacement with either the S605del or D316G POLD1 mutants (Fig. 2B). While reconstitution of POLD1-depleted cells with WT POLD1 restored translocation frequency, neither the S605del nor the D316G POLD1 mutants restored translocation frequency. Complementation with these mutants also had no effect on cell cycle distribution (Fig. 2C). These observations (summarized in SI Appendix, Table S6) are consistent with reports in yeast that Pol δ can promote 3′ end processing, microhomology-mediated end-joining, and translocations (31, 41).
POL δ Orchestrates Translocations by Alt-NHEJ.
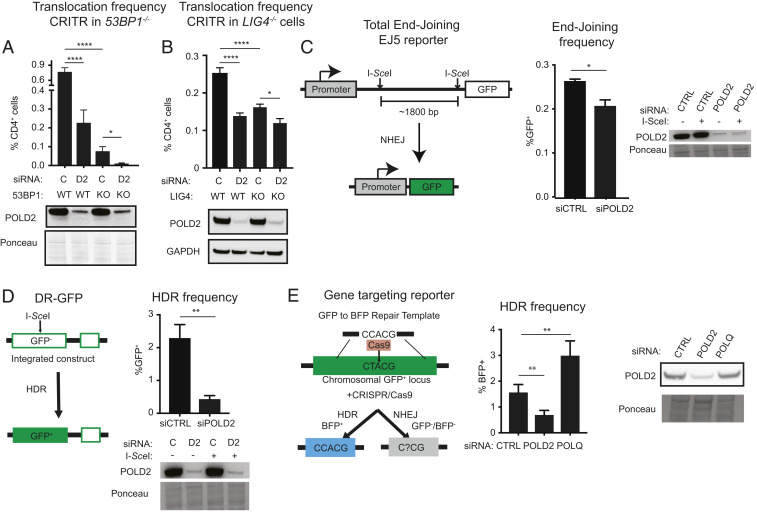
Because translocations are predominantly repaired using end-joining pathways in mammalian cells (22, 23, 28, 29), we hypothesized that Pol δ may play a role in Alt-NHEJ. As we previously reported (30), both LIG4−/− 293T cells and 53BP1−/− 293T cells had reduced translocation frequencies using the CRITR assay (Fig. 3 A and B). POLD2 depletion in 53BP1−/− 293T cells eliminated ∼80% of residual translocations (Fig. 3A), indicating that POLD2 can promote 53BP1-independent translocations. POLD2 depletion in LIG4−/− cells phenocopied POLD2 depletion in WT cells.
Fig. 3.
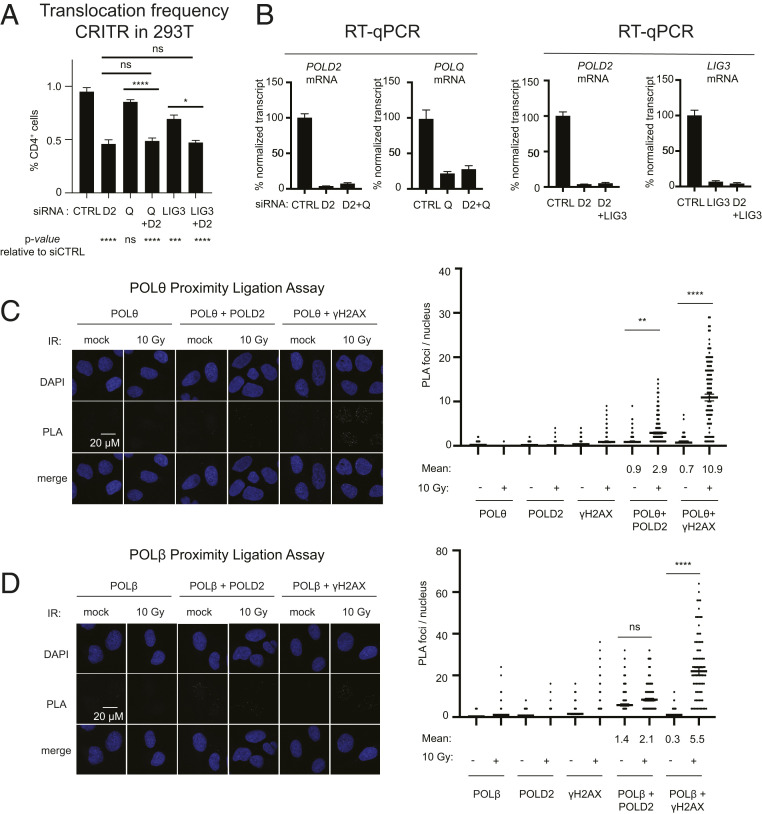
Pol δ participates in multiple DSB repair pathways. (A) Translocation frequency using CRITR and immunoblot after POLD2 depletion in WT and 53BP1−/− (knockout, KO) 293T cells. (B) Translocation frequency using CRITR and immunoblot after POLD2 depletion in WT and LIG4−/− (KO) 293T cells. (C) Schematic of the EJ5 end-joining reporter (43) in which two I-SceI–induced DSBs separated by ∼1,800 bp can be rejoined to juxtapose a promoter upstream of a GFP gene. Frequency of end-joining measured by EJ5 and immunoblot in HeLa cells transfected with the indicated siRNA (control nontargeting siRNA, siCTRL; siPOLD2, siPOLD2 #1) and expressing I-SceI. (D) Schematic of DR-GFP (48), frequency of HDR and immunoblot in U2OS DR-GFP cells transfected with siCTRL or siPOLD2 #1. (E) Schematic of gene targeting reporter (49), frequency of gene targeting, and immunoblot in 293T GFP cells transfected with the indicated siRNA, Cas9, gRNA, and the repair template. Data are presented as mean ± SE of n = 3. P values calculated using a one-way ANOVA with Tukey’s correction. *P < 0.05, **P < 0.01, ****P < 0.0001. Quantification of POLQ depletion with siRNA is shown in Fig. 6B.
To test the effects of POLD2 depletion on intrachromosomal repair, we first used a reporter that measures NHEJ between two I-SceI–induced DSBs spaced 1.8 kb apart (Fig. 3C) (42, 43). POLD2 depletion reduced NHEJ by only 20%, suggesting that this form of intrachromosomal NHEJ is less dependent on POLD2 compared with translocations. Based on reports that Pol δ promotes a key step in HDR in yeast (44–47), we next examined the contribution of POLD2 to HDR in human cells. To test this, we measured HDR frequency using two assays: 1) The DR-GFP reporter (48), in which the homologous template is 2.7 kb downstream of the DSB (Fig. 3D); and 2) a gene targeting reporter in which the homologous template is provided on a plasmid (Fig. 3E) (49). In both contexts, HDR was significantly decreased upon POLD2 depletion (Fig. 3 D and E). These findings are consistent with a recent report that depletion of other Pol δ subunits decreases HDR in human cells (36). Like POLD2, POL θ also promotes translocations through Alt-NHEJ (17, 50) but, in contrast with POLD2, depletion of POLQ increased HDR frequency (Fig. 3E).
POL δ Binds DNA DSBs in Proximity to γH2AX.
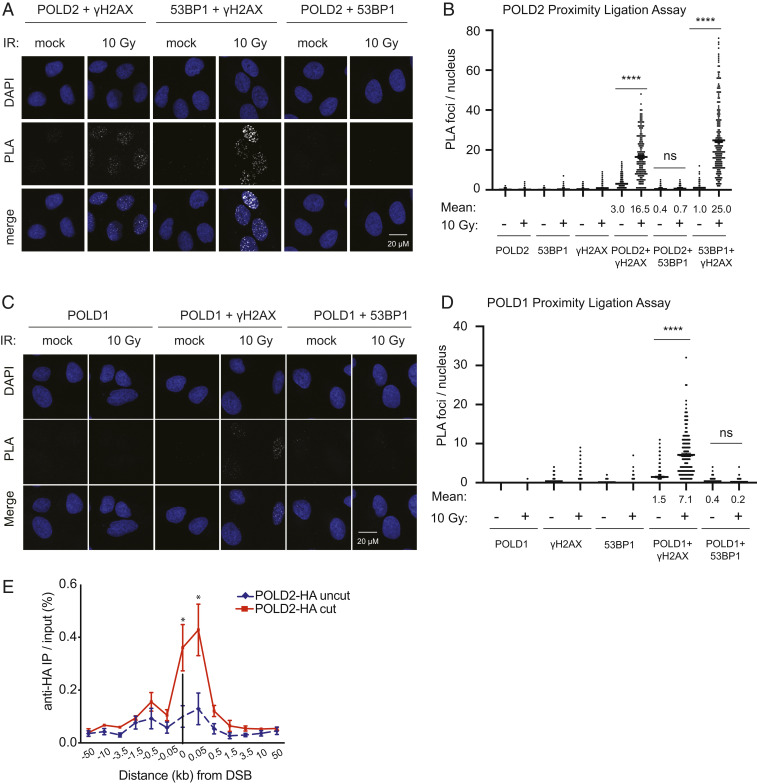
Pol δ is recruited to UV light-induced DNA damage in human cells and HO endonuclease-induced DSBs in budding yeast (31, 51, 52). Thus, we hypothesized that Pol δ is recruited directly to DSBs in human cells. Using PLAs, we monitored the interaction of POLD2 and POLD1 and γH2AX after 10 Gy IR in U2OS cells. Both POLD2 and POLD1 colocalized with γH2AX following IR treatment, albeit at lower frequencies than 53BP1-γH2AX colocalization (Fig. 4 A–D and SI Appendix, Fig. S2). Of note, we did not detect colocalization of either POLD2 or POLD1 with 53BP1 following IR (Fig. 4 A–D), which is consistent with our observation that POLD2 promotes translocations in 53BP1−/− cells (Fig. 3A). Using an orthogonal strategy, we asked whether POLD2 localizes to a single, targeted DSB. We introduced a single DSB with ZFNs in cells expressing HA-tagged POLD2. Chromatin immunoprecipitation (ChIP) for HA-POLD2 followed by qPCR demonstrated enrichment of sequences flanking the DSB site (Fig. 4E).
Fig. 4.
POLD localizes to DSBs. (A and B) PLA with antibodies against POLD2 and the indicated targets at 2 h following 10 Gy IR or mock treatment. Representative PLA panels (A) and quantification of PLA foci (B). (C and D) PLA with antibodies against POLD1 and the indicated targets as in A and B. More than or equal to 90 cells were imaged per condition per replicate. (E) HA-POLD2 occupancy at the AAVS1 locus 18 h after transient expression of ZFN by ChIP and quantitative PCR. Uncut, mock transfection; cut, AAVS1 ZFN. Data are presented as mean ± SE of n ≥ 2 independent experiments each performed in triplicate. P values for PLA calculated using a one-way ANOVA with Tukey’s correction. P values for ChIP calculated using Student’s t test, n = 4. *P < 0.05, ****P < 0.0001.
POL δ Promotes Sequence Modification at DSB Repair Junctions.
To define the effects of POLD2 on junction phenotypes, we cloned and sequenced individual translocation junctions from our CRITR assay. We noted that translocation junctions without any end processing (referred to as “exact”) represented a significantly higher fraction of translocations in POLD2-depleted cells compared to WT cells (Fig. 5A). Exact translocations occurred at a similar absolute frequency in POLD2-depleted and control cells (Fig. 5B); this suggests that exact junctions are POLD2-independent, whereas most junctions with sequence modifications were POLD2-dependent.
Fig. 5.
POLD2 promotes inaccurate NHEJ. (A) Relative proportions of repair classes in CRITR CD4/CD71 translocation junctions in 293T cells transfected with nontargeting siRNA (siCTRL) or siPOLD2 #1. Exact, no sequence loss or gain; Deletion, only sequence losses; Insertion, only sequence gains; Complex, deletions coupled with insertions. (B) Absolute frequencies of modified and exact translocation junction sequences. Modified sequences include deletions, insertions, and complex. (C) Insertion lengths at CRITR CD4/CD71 translocation junctions in 293T cells. Insertion lengths include insertions found in complex junctions. (D) Schematic of deep-sequencing assay coupled with Hi-FIBR analysis of junctions at a single DSB. (E) DSB repair classes and (F) microhomology usage for intrachromosomal repair of CRISPR/Cas9-induced DSB in the CD4 locus in 293T cells transduced with siRNA-resistant POLD2 or control. Cells were transfected with the indicated siRNA followed by Cas9 and gRNA targeting CD4 48 h later and DNA was harvested 48 h after CRISPR transfection. (G and H) Repair classes and microhomology usage for intrachromosomal repair of CRISPR/Cas9-induced DSB in the ESR1 locus in 293T cells as in E and F. Data are presented as mean ± SE of n = 3 independent experiments. P values calculated using a two-way ANOVA with Tukey’s correction (A and E–H) or Student’s t test (C). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Among translocation junctions with sequence modifications, insertions were shorter among POLD2-depleted cells than in controls cells (mean 54.6 bp vs. 26.2 bp; P < 0.0001) (Fig. 5C). Insertions that were long enough to align with known sequences (i.e., ≥20 bp) were predominantly templated from the Cas9 expression vector (SI Appendix, Fig. S3A). There were no differences among junctions with sequence modification in deletion length or microhomology usage between POLD2-depleted and control cells (SI Appendix, Fig. S3 B and C).
Next, we monitored intrachromosomal repair using targeted Cas9-mediated DSBs at either the CD4 or the ESR1 locus. Repair products were collected after 48 h, amplified in the linear range (SI Appendix, Fig. S4), and deep sequenced (total reads 30,928 to 121,372 per replicate) (SI Appendix, Tables S1 and S2). Reads were analyzed using Hi-FiBR (34), a bioinformatic approach for mapping individual reads to a predicted junction and characterizing the repair outcome (Fig. 5D and SI Appendix, Tables S3 and S4). As we cannot distinguish uncut loci from error-free repair, we focused our analysis on misrepaired junctions, similar to other recent studies (50, 53, 54). POLD2 depletion significantly reduced the proportion of junctions that contained deletions and increased the proportion containing insertions at the CD4 locus (Fig. 5E). Of note, the fraction of deletions with 0 bp of microhomology was significantly increased and those with ≥3 bp were decreased in POLD2-depleted cells, consistent with POLD2 participating in microhomology-mediated junctions (Fig. 5F). These effects were reversed by coexpression of siRNA-resistant POLD2. Depletion of POLD2 also led to reduced deletions, increased insertions and the use of longer microhomology at intrachromosomal repair junctions after a single DSB at the ESR1 locus (Fig. 5 G and H).
POLD1-depleted cells also had a reduced fraction of intrachromosomal repair junctions at the CD4 locus that harbored deletions (SI Appendix, Fig. S5A). The effect was reversed by reexpression of either WT POLD1 or POLD1 D316G but not by POLD1 S605del, which lacks DNA synthesis activity (SI Appendix, Fig. S5A). We also noted a small but statistically significant increase in the fraction of events with 0 bp of microhomology and decrease in the fraction with ≥3 bp of microhomology with POLD1 depletion (SI Appendix, Fig. S5B). This was reversed by coexpression of WT POLD1 but not by either mutant. We did not observe any consistent changes in length of deletions or insertions from depletion of POLD1 or POLD2 (SI Appendix, Fig. S5 C–F).
POL δ Participates in Alt-NHEJ with Features Distinct from POL θ- or LIG3-Dependent Alt-NHEJ.
To investigate additional factors that may contribute to Pol δ-dependent translocation formation, we measured functional interactions of POLD2 with multiple factors (17, 21, 29, 50). We assumed that depletion conditions that maintained cell doubling rate at >90% of control per cell cycle were unlikely to be associated with marked secondary effects on replication or cell stress. Thus, we measured proliferation after knockdown of single factors or combinations at 5 d (or approximately five doublings of transfected 293T cells) and further investigated conditions where proliferation was >59% of control (0.905). Codepletion of POLQ and POLD2 resulted in >40% reduction in cell growth at day 5 (SI Appendix, Fig. S6A). In addition, POLQ-depletion did not elicit a significant change in translocation frequency in our CRITR assay (Fig. 6 A and B). Therefore, we could not infer a genetic relationship between Pol θ and POLD2 from these data without the possibility for significant confounding effects from codepletion. We observed a modest colocalization between Pol θ and POLD2 using PLA (Fig. 6C). In contrast to POLQ, depletion of LIG3 led to a significant decrease in the frequency of translocations (Fig. 6 A and B). Similar to LIG4 knockout (Fig. 3B), codepletion of LIG3 and POLD2 phenocopied depletion of POLD2 alone, suggesting that a subset of POLD2-dependent translocations involve LIG3. Based on a report that Pol β participates in Alt-NHEJ (27), we also depleted Pol β. Approximately 75% knockdown of Pol β resulted in ∼50% inhibition of cell growth at day 5 (SI Appendix, Fig. S6 A and B), precluding analysis of its role in translocations. PLA analysis between Pol β and POLD2 showed no significant IR-induced colocalization of the two factors (Fig. 6D).
Fig. 6.
Interactions between POLD2 and Alt-NHEJ factors in translocation formation. (A and B) Translocation frequency in CRITR assay (30) using 293T cells transfected with siCTRL or siRNA targeting POLD2 (siPOLD2 #1, D2), POLQ (Q), LIG3 or combinations, and 72 h later transfected with the same siRNAs and Cas9/gRNA. Cells were collected after 48 h for flow cytometry (A) and RT-qPCR for the indicated transcripts (B). Data are presented as mean ± SE of n = 3 independent experiments. (C) PLA with antibodies against POLθ and the indicated targets at 2 h following 10 Gy IR or mock treatment. Representative PLA panels and quantification of PLA foci. (D) PLA with antibodies against POLβ and the indicated targets as in C. Greater than or equal to 90 cells were imaged per condition per replicate and data are presented as mean ± SE of n ≥ 2 independent experiments. P values calculated using a one-way ANOVA with Tukey’s correction. *P < 0.05, **P < 0.01, ****P < 0.0001.
In the Hi-FIBR analysis of intrachromosomal repair at the CD4 locus, depletion of LIG3 resulted in a significant increase in the proportion of junctions that contain deletions and a decrease in the junctions that contain insertions. In contrast, depletion of POLD2 led to a significant decrease of junctions with deletions and an increase in those containing insertions (SI Appendix, Fig. S6C). We observed a similar pattern of opposite effects for depletion of LIG3 versus depletion of POLD2 in microhomology usage: LIG3 depletion decreased the fraction of events with 0 bp of microhomology and increased the fraction with ≥3 bp of microhomology, while POLD2 depletion had the opposite effects (SI Appendix, Fig. S6D). Similar to siPOLD2 and consistent with previous reports (17, 50), Pol θ depletion led to a significant decrease in junctions with ≥3 bp of microhomology (SI Appendix, Fig. S6D).
Discussion
Here we identified Pol δ subunits as translocation-promoting factors across multiple human cell lines, independent of whether the DSBs were introduced by ZFNs or CRISPR/CAS9. To confirm that translocation formation was not impacted by alterations induced by POLD2 knockdown (i.e., secondary effects), we monitored cell cycle status, replication stress levels, and differential gene expression in POLD2-depleted cells. The absence of secondary effects in these assays suggests that POLD2 promotes translocations by directly affecting DSB repair rather than indirectly by affecting other cellular processes. A combination of control experiments (i.e., cutting efficiency, endonuclease expression, transcriptional profiling, cell cycle analysis, and assays to identify induction of stress or other response pathways germane to the factor being assessed) is important to ensure that confounding effects are minimized when assaying genetic manipulations of DNA repair factors.
The absence of effects from POLD2 knockdown on cell cycle and replication stress seems counterintuitive, as depletion should decrease origin firing or lead to more collapsed replication forks. However, previous studies reported that depletion of POLD3 or POLD4 did not lead to proliferative defects in U2OS or HeLa cells (36, 37). These data suggest that depletion of Pol δ structural subunits does not impair proliferation, perhaps due to residual amounts of Pol δ after depletion being sufficient for DNA replication (55). Indeed, DNA fiber assays did not indicate any changes in replication fork speed in POLD3- and POLD1-depleted cells, although interorigin distance was longer, presumably due to decreased origin firing (36).
Our data suggest that Pol δ subunits promote Alt-NHEJ at both intra- and interchromosomal junctions in human cells, similar to their homologs in yeast (31, 56, 57). In human cells, translocation junctions are most consistent with repair by some form of NHEJ that may involve resection to single-strand tails and gap-filling by polymerases. Pol μ, λ, β, and θ can all perform gap-filling synthesis during the final steps of end-joining in human cells (27, 58, 59) and large-scale complex rearrangements commonly have signatures of polymerase involvement (22, 23, 28, 29, 60–62). We noted that POLD2 plays a role in HDR (Fig. 3 C–E), a repair process that also requires DNA synthesis. The similar effects of POLD2 and POLD1 knockdown on translocation frequency (Figs. 1 and 2) and the inability of POLD1 synthesis and proof-reading mutants to complement POLD1 knockdown (Fig. 2B) indicate that Pol δ enzymatic activity is involved in these translocations.
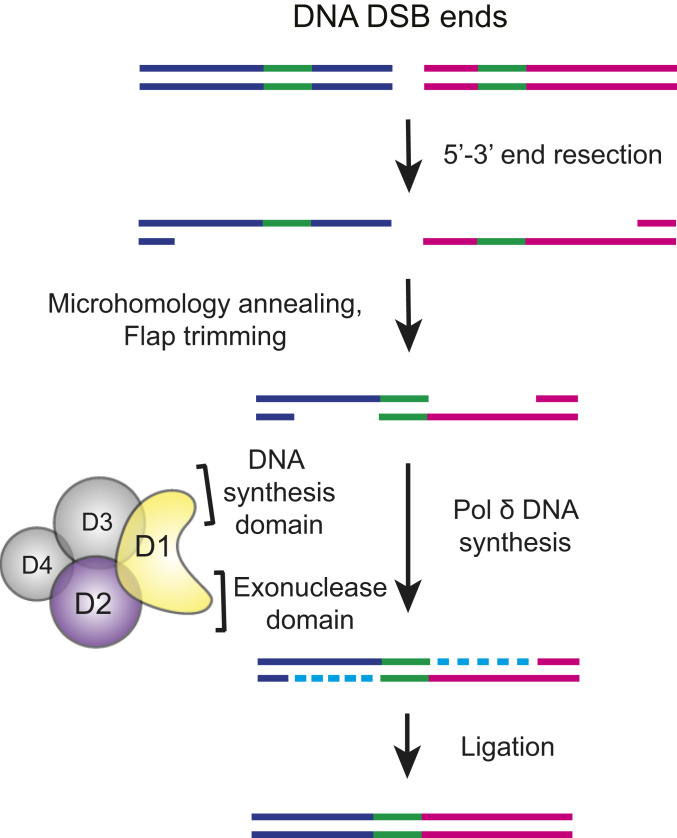
Based on our data and the known functions of Pol δ, we propose a model in which Pol δ promotes the extension of annealed microhomologies that are revealed by processing of DSBs to single-strand intermediates (Fig. 7). This model is similar to ones proposed for Pol θ and Pol β action during DSB repair (17, 27). Together, the data suggest that multiple polymerases may 1) serve redundant functions, 2) be required for distinct steps of Alt-NHEJ, and 3) act on distinct intermediates generated during Alt-NHEJ. Whether there are specific intermediates that favor one polymerase over another or there is a requirement for multiple polymerases in a specified sequence has not been clarified. For example, Pol θ and Pol β may initiate DNA synthesis from microhomologies, which is followed by Pol δ recruitment and more extensive DNA synthesis. Indeed, several studies have demonstrated or suggested that Pol δ and Pol θ can iteratively bind, extend, and dissociate from DNA (63, 64). The differential effects of POLD2, LIG3 and Pol θ depletion (alone or in combinations) on junction phenotypes (summarized in SI Appendix, Table S7) underscore the complex interplay of these factors.
Fig. 7.
Model for Pol δ involvement in Alt-NHEJ. Model for involvement of Pol δ in DSB repair that leads to imprecise junctions. Pol δ promotes Alt-NHEJ by catalyzing DNA synthesis after DSB end processing and annealing of sequences with microhomology. The role of POLD4 remains unclear as does the specific requirement for individual ligases.
Specific to translocations, we found that depletion of POLD2 was additive with 53BP1 knockout, with the combination of POLD2 and 53BP1 leading to nearly undetectable levels of translocations in our CRITR assay. One interpretation of these data is that translocation events depend on either 53BP1 or POLD2. In contrast, POLD2 depletion in LIG4−/− or LIG3-depleted cells did not further reduce translocation frequency compared with POLD2 depletion in WT cells. This suggests that POLD2 is upstream of these ligases, which may serve redundant functions. That hypothesis is consistent with previous data that XRCC4/LIG4 efficiently ligates ends paired through microhomology in the absence of Ku, and thus could complete repair events that involve Alt-NHEJ factors (65).
Multiple aspects of Pol δ’s role in DSB repair require further study. First, POLD2 and POLD3 also associate with Pol ζ subunits, implying that POLD2 depletion could impair Pol ζ function (66–68). However, we report that depletion of POLD1 (which is not part of Pol ζ) phenocopies POLD2 depletion in reducing translocation frequency and repair junction characteristics. Furthermore, we demonstrate that the DNA synthesis and proof-reading activities of POLD1 are important for these phenotypes, which directly implicates Pol δ. In addition, previous studies in budding yeast have shown that mutations in rev3, a subunit of Pol ζ, do not result in changes in translocation frequency or inaccurate end-joining, suggesting that Pol ζ does not modulate translocation frequency in yeast (31, 41). Taken together, these findings support a role for Pol δ in translocation formation and error-prone repair at DSBs. While these data do not rule out the possibility that Pol ζ also plays a role in the phenotypes observed with POLD2 depletion, it suggests the role if any is relatively minor.
Second, we determined that POLD2 is recruited to both endonuclease- and IR-induced DSBs in human cells but, based on our PLA, the recruitment appeared to be less robust than recruitment of 53BP1. This could be an artifact of the PLA and the different antibodies used to detect each protein. Alternatively, POLD2 may be recruited to only a subset of DSBs, for example those involving DSB resection. A third possibility is that the dwell time of POLD2 at a DSB may be short and thus we would only see a limited number of POLD2/γ-H2AX interactions at any given time. Using ChIP at a ZFN-induced DSB in 293T cells, POLD2 was most enriched within a short distance of the DSB site. This could indicate that Pol δ is primarily involved in events with short sequence modifications. We note that both the CRITR and ZiTR assays are unable to detect repair events that occur following large deletions, although these events are likely to be rare. Alternatively, Pol δ could be primarily involved in modifying end sequences early during the repair (e.g., through its exonuclease domain) or very late in the process. A third possibility is more prosaic but possibly more likely: Specifically, that events where Pol δ functions farther away from the DSB are simply diluted out (i.e., fewer molecules of Pol δ per kilobase of sequence) and thus appear to be unenriched for Pol δ.
Third, the requirements for Pol δ recruitment to DSBs are currently unknown. In vitro experiments using double Holliday junction substrates suggest that a double-strand to single-strand transition is sufficient to recruit Pol δ (69). Immunofluorescence studies in human cells have revealed that depletion of RFC140 leads to >50% reduction in POLD1 recruitment to XPA laser strips following UV damage (51, 70, 71). Curiously, recruitment of the Pol δ processivity factor proliferating cell nuclear antigen (PCNA) is not affected by RFC140 depletion, suggesting that POLD1 recruitment could be independent of PCNA recruitment and loading in some contexts (71). As such, the requirement for PCNA recruitment in Alt-NHEJ events involving Pol δ remains to be clarified. Finally, any contribution of the POLD4 subunit to Alt-NHEJ events including translocations has not been determined.
We noticed that following POLD2 depletion these profiles shifted, resulting in a decrease in the relative fraction of simple deletions following repair. Additionally, we found that deletions in a POLD2-deficient setting exhibited significantly less microhomology than WT counterparts. These findings are concordant with a study in budding yeast showing that Alt-NHEJ is decreased in Pol3 temperature-sensitive and Pol32 mutants, although they did not examine microhomology usage (41, 57, 72). Because POLD2 depletion can affect multiple pathways of DSB repair, including HDR and NHEJ, the phenotypes observed at single DSB cannot be clearly attributed to its effects on a single repair mechanism.
Supplementary Material
Acknowledgments
We thank Steve Elledge, Ralph Scully, and members of the D.M.W. laboratory for thoughtful comments. This work was supported by a National Science Foundation Graduate Research Fellowship DGE1144152 (to J.V.L.), NCI R35 CA239158 (to D.M.W.), and an NIA K01 AG056554 (to T.A.D.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014176117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Zhang Y., Rowley J. D., Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair 5, 1282–1297 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Roukos V., Misteli T., The biogenesis of chromosome translocations. Nat. Cell Biol. 16, 293–300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aplan P. D., Causes of oncogenic chromosomal translocation. Trends Genet. 22, 46–55 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sallmyr A., Tomkinson A. E., Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J. Biol. Chem. 293, 10536–10546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciccia A., Elledge S. J., The DNA damage response: Making it safe to play with knives. Mol. Cell 40, 179–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McVey M., Khodaverdian V. Y., Meyer D., Cerqueira P. G., Heyer W. D., Eukaryotic DNA polymerases in homologous recombination. Annu. Rev. Genet. 50, 393–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright W. D., Shah S. S., Heyer W. D., Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 293, 10524–10535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang H. H. Y., Pannunzio N. R., Adachi N., Lieber M. R., Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang H. H., Lieber M. R., Structure-specific nuclease activities of Artemis and the Artemis: DNA-PKcs complex. Nucleic Acids Res. 44, 4991–4997 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y.et al., A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell 16, 701–713 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Grawunder U.et al., Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388, 492–495 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Iliakis G., Murmann T., Soni A., Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 793, 166–175 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Stephens P. J.et al., Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemée F.et al., DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl. Acad. Sci. U.S.A. 107, 13390–13395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boboila C.et al., Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J. Exp. Med. 207, 417–427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansour W. Y., Rhein T., Dahm-Daphi J., The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 38, 6065–6077 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mateos-Gomez P. A.et al., Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahrabi S.et al., A role for human homologous recombination factors in suppressing microhomology-mediated end joining. Nucleic Acids Res. 44, 5743–5757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deriano L., Stracker T. H., Baker A., Petrini J. H., Roth D. B., Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol. Cell 34, 13–25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masani S., Han L., Meek K., Yu K., Redundant function of DNA ligase 1 and 3 in alternative end-joining during immunoglobulin class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 113, 1261–1266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu G.et al., Ligase I and ligase III mediate the DNA double-strand break ligation in alternative end-joining. Proc. Natl. Acad. Sci. U.S.A. 113, 1256–1260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Jasin M., An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat. Struct. Mol. Biol. 18, 80–84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghezraoui H.et al., Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell 55, 829–842 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma S.et al., Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis. 6, e1697 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvajal-Garcia J.et al., Mechanistic basis for microhomology identification and genome scarring by polymerase theta. Proc. Natl. Acad. Sci. U.S.A. 117, 8476–8485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yousefzadeh M. J.et al., Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 10, e1004654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray S.et al., DNA polymerase beta participates in DNA end-joining. Nucleic Acids Res. 46, 242–255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinstock D. M., Brunet E., Jasin M., Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat. Cell Biol. 9, 978–981 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simsek D.et al., DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 7, e1002080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Day T. A.et al., PARP3 is a promoter of chromosomal rearrangements and limits G4 DNA. Nat. Commun. 8, 15110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer D., Fu B. X., Heyer W. D., DNA polymerases δ and λ cooperate in repairing double-strand breaks by microhomology-mediated end-joining in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 112, E6907–E6916 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y.et al., Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell 48, 723–733 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown A. J.et al., High-throughput analysis of DNA break-induced chromosome rearrangements by amplicon sequencing. Methods Enzymol. 601, 111–144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.You Z., Bailis J. M., DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 20, 402–409 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tumini E., Barroso S., -Calero C. P., Aguilera A., Roles of human POLD1 and POLD3 in genome stability. Sci. Rep. 6, 38873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costantino L.et al., Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 343, 88–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng D. Q., Petes T. D., Genome instability induced by low levels of replicative DNA polymerases in yeast. Genes 9, 539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weedon M. N.et al., An in-frame deletion at the polymerase active site of POLD1 causes a multisystem disorder with lipodystrophy. Nat. Genet. 45, 947–950 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellido F.et al., POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: Review of reported cases and recommendations for genetic testing and surveillance. Genet. Med. 18, 325–332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tseng S. F., Gabriel A., Teng S. C., Proofreading activity of DNA polymerase Pol2 mediates 3′-end processing during nonhomologous end joining in yeast. PLoS Genet. 4, e1000060 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gunn A., Stark J. M., I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 920, 379–391 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Bennardo N., Cheng A., Huang N., Stark J. M., Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holmes A. M., Haber J. E., Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96, 415–424 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Lydeard J. R., Jain S., Yamaguchi M., Haber J. E., Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448, 820–823 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Maloisel L., Fabre F., Gangloff S., DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol. Cell. Biol. 28, 1373–1382 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.House N. C.et al., Distinct roles for S. cerevisiae H2A copies in recombination and repeat stability, with a role for H2A.1 threonine 126. eLife 8, e53362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esashi F.et al., CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 434, 598–604 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Glaser A., McColl B., Vadolas J., GFP to BFP conversion: A versatile assay for the quantification of CRISPR/Cas9-mediated genome editing. Mol. Ther. Nucleic Acids 5, e334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyatt D. W.et al., Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell 63, 662–673 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hashiguchi K., Matsumoto Y., Yasui A., Recruitment of DNA repair synthesis machinery to sites of DNA damage/repair in living human cells. Nucleic Acids Res. 35, 2913–2923 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chea J.et al., Spatiotemporal recruitment of human DNA polymerase delta to sites of UV damage. Cell Cycle 11, 2885–2895 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khodaverdian V. Y.et al., Secondary structure forming sequences drive SD-MMEJ repair of DNA double-strand breaks. Nucleic Acids Res. 45, 12848–12861 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Layer J. V.et al., Parp3 promotes long-range end joining in murine cells. Proc. Natl. Acad. Sci. U.S.A. 115, 10076–10081 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J.et al., The second subunit of DNA polymerase delta is required for genomic stability and epigenetic regulation. Plant Physiol. 171, 1192–1208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee K., Lee S. E., Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics 176, 2003–2014 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Villarreal D. D.et al., Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet. 8, e1003026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pryor J. M.et al., Essential role for polymerase specialization in cellular nonhomologous end joining. Proc. Natl. Acad. Sci. U.S.A. 112, E4537–E4545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kent T., Chandramouly G., McDevitt S. M., Ozdemir A. Y., Pomerantz R. T., Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol. 22, 230–237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang C. Z.et al., Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crasta K.et al., DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richardson C., Moynahan M. E., Jasin M., Double-strand break repair by interchromosomal recombination: Suppression of chromosomal translocations. Genes Dev. 12, 3831–3842 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hedglin M., Pandey B., Benkovic S. J., Stability of the human polymerase δ holoenzyme and its implications in lagging strand DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 113, E1777–E1786 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beagan K.et al., Drosophila DNA polymerase theta utilizes both helicase-like and polymerase domains during microhomology-mediated end joining and interstrand crosslink repair. PLoS Genet. 13, e1006813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu J.et al., XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 26, 1010–1023 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Makarova A. V., Stodola J. L., Burgers P. M., A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 40, 11618–11626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson R. E., Prakash L., Prakash S., Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc. Natl. Acad. Sci. U.S.A. 109, 12455–12460 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siebler H. M., Lada A. G., Baranovskiy A. G., Tahirov T. H., Pavlov Y. I., A novel variant of DNA polymerase ζ, Rev3ΔC, highlights differential regulation of Pol32 as a subunit of polymerase δ versus ζ in Saccharomyces cerevisiae. DNA Repair 24, 138–149 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J., Holzschu D. L., Sugiyama T., PCNA is efficiently loaded on the DNA recombination intermediate to modulate polymerase δ, η, and ζ activities. Proc. Natl. Acad. Sci. U.S.A. 110, 7672–7677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Overmeer R. M.et al., Replication factor C recruits DNA polymerase delta to sites of nucleotide excision repair but is not required for PCNA recruitment. Mol. Cell. Biol. 30, 4828–4839 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ogi T.et al., Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell 37, 714–727 (2010). [DOI] [PubMed] [Google Scholar]
- 72.Galli A., Chan C. Y., Parfenova L., Cervelli T., Schiestl R. H., Requirement of POL3 and POL4 on non-homologous and microhomology-mediated end joining in rad50/xrs2 mutants of Saccharomyces cerevisiae. Mutagenesis 30, 841–849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.