Significance
Quantitative information about strengths of amide nitrogen−amide oxygen hydrogen bonds and π-system and hydrophobic interactions involving amide-context sp2 and/or sp3 carbons is needed to assess their contributions to specificity and stability of protein folds and assemblies in water, as well as to predict or interpret how urea and other amides interact with proteins and affect protein processes. Here we obtain this information from thermodynamic measurements of interactions between amide compounds in water and an analysis that determines intrinsic strengths of atom−atom interactions, relative to water and per unit area of each atom type present. These findings allow prediction or interpretation of effects of any amide on protein processes from structure, and may be useful to analyze protein interfaces.
Keywords: preferential interactions, amide atom interactions, aqueous interactions, additivity, thermodynamics
Abstract
Folding and other protein self-assembly processes are driven by favorable interactions between O, N, and C unified atoms of the polypeptide backbone and side chains. These processes are perturbed by solutes that interact with these atoms differently than water does. Amide NH···O=C hydrogen bonding and various π-system interactions have been better characterized structurally or by simulations than experimentally in water, and unfavorable interactions are relatively uncharacterized. To address this situation, we previously quantified interactions of alkyl ureas with amide and aromatic compounds, relative to interactions with water. Analysis yielded strengths of interaction of each alkylurea with unit areas of different hybridization states of unified O, N, and C atoms of amide and aromatic compounds. Here, by osmometry, we quantify interactions of 10 pairs of amides selected to complete this dataset. An analysis yields intrinsic strengths of six favorable and four unfavorable atom−atom interactions, expressed per unit area of each atom and relative to interactions with water. The most favorable interactions are sp2O−sp2C (lone pair−π, presumably n−π*), sp2C−sp2C (π−π and/or hydrophobic), sp2O−sp2N (hydrogen bonding) and sp3C−sp2C (CH−π and/or hydrophobic). Interactions of sp3C with itself (hydrophobic) and with sp2N are modestly favorable, while sp2N interactions with sp2N and with amide/aromatic sp2C are modestly unfavorable. Amide sp2O−sp2O interactions and sp2O−sp3C interactions are more unfavorable, indicating the preference of amide sp2O to interact with water. These intrinsic interaction strengths are used to predict interactions of amides with proteins and chemical effects of amides (including urea, N-ethylpyrrolidone [NEP], and polyvinylpyrrolidone [PVP]) on protein stability.
Biopolymer self-assembly in water, including folding, binding, droplet formation, phase separation, and formation of the functional protein and nucleic acid complexes of the cell, is driven by net-favorable interactions between C, N, and O unified atoms of their biochemical functional groups, relative to interactions with water. To understand the energetics of these processes and relate thermodynamic and structural information, strengths of interaction of the different types of C, N, and O unified atoms with one another, relative to their interactions with water, must be determined. Effects of biochemical solutes and noncoulombic effects of salts from the Hofmeister series on these biopolymer processes result from preferential interactions of the C, N, and O atoms of the solute (and inorganic ions of the salt) with the C, N, and O atoms of the biomolecule (1, 2). Quantitative information about preferential interactions of the various types of C, N, and O atoms of biomolecules and solutes will therefore be useful in analyzing both self-assembly interactions and solute effects on these interactions.
Hydrogen bonding between amide sp2O and sp2N unified atoms (3–6) and the hydrophobic effect of reducing the exposure of sp2C and sp3C atoms to water (7–10) have long been recognized to be key determinants of specificity and stability of protein assemblies and complexes. In addition, n−π* interactions [a type of lone pair (lp)–π interaction (11)] between amide sp2O and amide or aromatic sp2C (12–17), π−π interactions of sp2C with sp2C (18, 19) and CH−π interactions of sp3C with sp2C (20–23) have been characterized by structural, spectroscopic, and computational studies. Much less is known about the relative strengths of these and other amide atom−atom contacts in water, including interactions of amide sp2N with amide sp2N, sp3C, and sp2C and interactions of amide sp2O with amide sp2O and sp3C.
Preferential interactions of biochemical solutes and Hofmeister salts with other solutes or biopolymers, relative to interactions with water, are quantified by chemical potential derivatives where the subscripts “2” and “3” refer to the two solutes and µ23 = µ32 (1, 2). These µ23 values, related to transfer free energies, are determined by osmometry or solubility assays (20, 24–32). Integration of the radial distribution (33–36) of one solute in the vicinity of the other, obtained from molecular dynamics simulations (36–41), also yields µ23 (31).
Experimental research and analysis extending over the last decade (24–32) has shown that µ23 values are accurately described as a sum of contributions from interactions of solute 3 with the different types (hybridization states) of C, N, and O atoms on solute 2,
| [1] |
The choice of which solute to designate as component 2 or 3 is arbitrary because µ23 = µ32.
In Eq. 1, each intensive quantity is a thermodynamic coefficient (called a one-way alpha value) that quantifies the strength of interaction of the amide compound designated component 3 (e.g., urea) with a unit area of one of the i different types of C, N, and O atoms on the set of amide compounds (each designated component 2), relative to interactions with water. The extensive quantity in Eq. 1 is the water-accessible surface area (abbreviated ASA, in units of angstrom squared [Å2] where 1 Å = 0.1 nm), of the ith atom type of the amide solute (component 2) whose interaction with amide solute 3 is quantified by . Examples of Eq. 1 for the interaction of two amide compounds investigated here are provided in SI Appendix, section III.
Eq. 1 is based on the two hypotheses that contributions to µ23 from different weak solute−atom interactions are additive and increase in proportion to the ASA of that C, N, or O atom. Additivity has been tested and validated by analysis of sets of µ23 values using Eq. 1 because the sizes of the µ23 datasets greatly exceed the number of one-way alpha values determined from the analysis. ASA is found to be a better choice of extensive variable than the number of atoms or weighted number of atoms (31, 32). Using one-way alpha values , effects of solutes (species 3) on biopolymer (species 2) processes are predicted or interpreted in terms of the interaction of solute 3 with the different types of biopolymer ASA exposed or buried in the process.
The µ23 values can be interpreted as free energy changes for transfer of a solute from a two-component solution to a three-component solution in which the concentration of the other solute is 1 molal. Originally, effects of urea and osmolytes on protein stability were interpreted assuming additivity of transfer free energy contributions from the peptide backbone and each of the nineteen different amino acid side chains that are exposed to the solution in unfolding (42–44). These 20 side chain and backbone transfer free energies were obtained from amino acid and dipeptide solubility data, also assuming additivity. Analysis of urea effects on protein stability using Eq. 1 involves many fewer parameters [from as few as two (45) to four (31) or seven (26, 27) one-way alpha values depending on the extent of coarse graining of the ASA exposed in unfolding]. One-way alpha values are interpretable in terms of the local accumulation or exclusion of the solute in the vicinity of a particular type of atom on the model compound or protein, using the solute partitioning model (2, 20, 24–29, 46, 47).
Trends in one-way alpha values for interactions of the series of alkyl ureas with the different atoms of amide compounds (31) indicate that these one-way alpha values can be dissected further. This analysis, reported below, yields a set of 10 two-way alpha values quantifying strengths of all pairwise interactions (favorable and unfavorable) between the different types of unified C, N, and O atoms of amide compounds (amide sp2O, sp2N, sp2C; aliphatic sp3C) relative to water and per unit ASA of each atom. No experimental information of this type was previously available. These two-way alpha values are also useful to predict strengths of interaction of other amide compounds and the effects of amide compounds on protein processes like folding in which most of the change in ASA is from amide and hydrocarbon atoms. Our previous study focused on the series of alkylated ureas, all of which have small water-accessible surface area of amide sp2C atoms. Here we extend the amide dataset by determining µ23 values for interactions of five other amides, including formamide and N-methylformamide, which have large amide sp2C ASA, and malonamide and N-acetylalanine N-methylamide (aama), which have two amide groups and correspondingly larger amide sp2O ASA.
Results
Analysis to Determine Atom−Atom Interactions from Solute−Atom Interactions.
For interactions of a series of urea and alkyl ureas (component 3) with amide compounds (component 2), analyzed by Eq. 1 as summarized above, each of the solute (3)−atom (i) one-way alpha values exhibited a regular progression with increasing alkylation (and reduced exposure of amide nitrogen) of the urea (SI Appendix, Fig. S1 and ref. 31). Motivated by this observation, here we test the hypothesis that each of these one-way alpha values can be dissected into additive, ASA-based contributions from the interaction of the different types of atoms on amide solute 3 (sp2O, N, C, and sp3C) with the ith type of atom (also sp2O, N, C, or sp3C) on amide solute 2.
| [2] |
Eq. 2 for the one-way alpha value is completely analogous to Eq. 1 for µ23. In Eq. 2, each intensive quantity is the strength of interaction of a unit area of atom j of solute 3 with a unit area of atom i of solute 2, and the corresponding extensive quantity is the accessible surface area of atom type j on solute 3. For the amide−amide interactions of interest here, an example with all of the individual terms in the sum in Eq. 2 is provided in SI Appendix, section III. The hypotheses of additivity and ASA dependence of the contributions to the one-way alpha value are tested concurrently with the determination of two-way alpha values , because the number of equations (like Eq. 2) greatly exceeds the number of unknowns being determined (see below). A direct but semiquantitative application of Eq. 2 to determine two-way alpha values from differences in one-way alpha values for amides differing primarily in ASA of one type of unified atom is given in SI Appendix, sections I and III and Eq. S1.
Combination of Eqs. 1 and 2 gives the proposed dissection of any solute−model compound µ23 values into contributions from the interactions of accessible atoms of the solute with accessible atoms of the model compound,
| [3] |
where . The complete set of terms in this double sum for the amide compounds investigated here is given in SI Appendix, section III.
As an interpretation of one term in Eq. 3, consider the contribution to µ23 from the interaction of amide sp2N of one amide solute (component 2) with amide sp2O on a second amide solute (component 3), relative to interactions with water, given by . The product of ASA values is proportional to the probability that a contact between the two solutes involves sp2N atom(s) of solute component 2 and sp2O atom(s) of solute component 3, and the two-way alpha value is the strength of that interaction per unit ASA of both atom types, again relative to water. These two-way alpha values are useful to predict or interpret µ23 values for interactions of amides for which one-way alpha values are not available, and to predict or interpret effects of these amides on protein processes in terms of structural information. Two-way alpha values may also be useful in analyses of atom−atom interactions in protein assemblies and in binding interfaces.
Vapor Pressure Osmometry Determinations of Interactions of Amides with Large ASA of Amide sp2C and sp2O.
Previously, we determined one-way alpha values (Eq. 1) quantifying interactions of urea and six alkyl urea solutes with the types of unified atoms of amide (sp2O, sp2N, sp2C; sp3C) and aromatic hydrocarbon (sp2C) compounds using osmometric and solubility studies (31). Trends in these one-way alpha values with increasing alkylation of the urea showed which atom−atom interactions are favorable and which are unfavorable. However, preliminary tests of Eqs. 2 and 3 using the 95 values from this previous study revealed that these were insufficient to accurately quantify all atom−atom interactions (two-way alpha values) involving amide sp2O and/or sp2C.
Here we determine values by osmometry for an additional 10 interactions of five amide compounds, including interactions of two amides with large ASA of amide sp2C (formamide, N-methylformamide) with one another and with two amides with large ASA of amide sp2O (malonamide, aama). Interactions of these four amides with propionamide are also determined. In addition to their significance for the two-way analysis proposed here, these measurements also permit the determination of one-way alpha values for the interactions of these five amides with amide sp2O, sp2N, sp2C, and aliphatic sp3C atoms, increasing the number of amide compounds for which one-way alpha values (Eq. 1) are available from 7 to 12.
For uncharged solutes at concentrations up to ∼1 molal, the difference ΔOsm = Osm(m2,m3) – Osm(m2) – Osm(m3) between the osmolality (Osm) of a three-component solution and the two corresponding two-component solutions is proportional to the product of solute molal concentrations (m2m3) with proportionality constant μ23/RT (31).
| [4] |
If ΔOsm is negative, μ23 is negative, μ2 decreases with increasing m3, and the interaction of the two solutes is favorable.
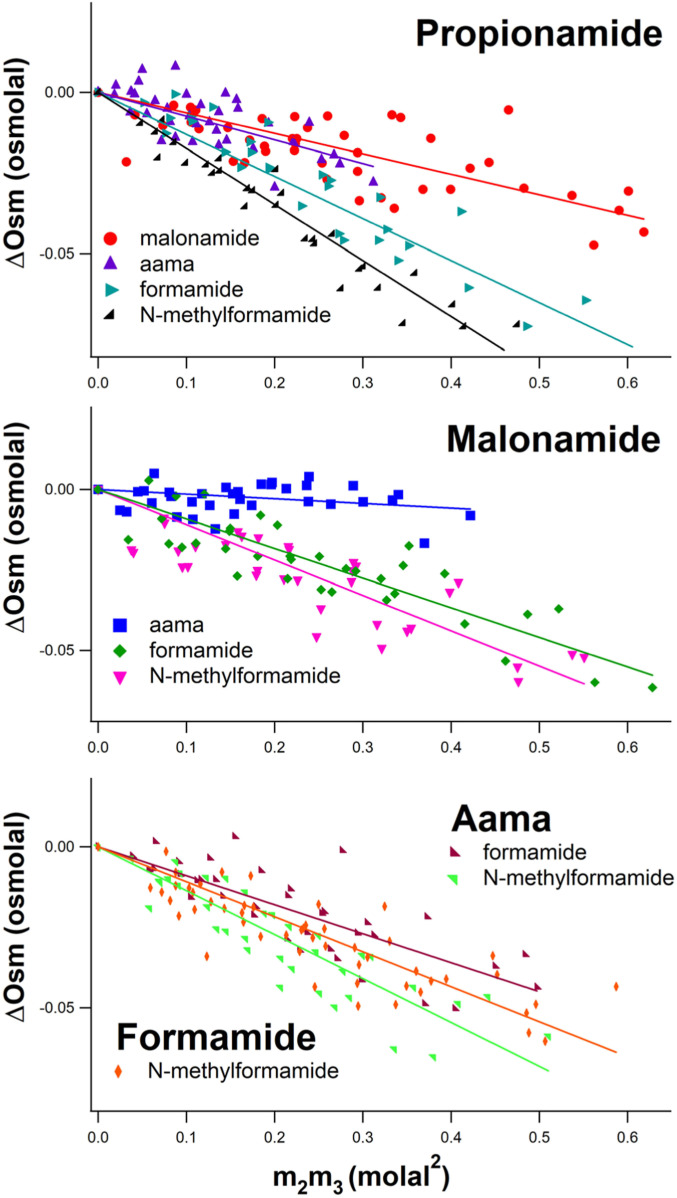
For each of the 10 pairs of amides investigated here, ΔOsm is plotted vs. m2m3 in Fig. 1. All µ23 values are negative, indicating favorable interactions between all 10 pairs of amides investigated here. Values of µ23 at 23 °C obtained from these slopes are listed in SI Appendix, Table S1. Of these, the interaction of malonamide and aama (Fig. 1, Middle) is the least favorable (µ23 = −8.6 ± 2.2 cal⋅mol−1⋅molal−1), and the interaction of propionamide and N-methylformamide (Fig. 1, Top) is the most favorable (µ23 = −102 ± 1.9 cal⋅mol−1⋅molal−1). Although there is substantial scatter in the data for some pairs of amides, slopes µ23/RT are quite well determined (SI Appendix, Table S1) because the intercept is constrained to be zero.
Fig. 1.
Osmometric determinations of preferential interactions of pairs of amide compounds in water. (Top) Propionamide-amide interactions. (Middle) Malonamide-amide interactions. (Bottom) N-acetylalanine N-methylamide (aama)-amide and formamide-amide interactions. Osmolality differences between a three-component solution of two amide compounds and the two corresponding two-component solutions, determined by VPO at 23 °C, are plotted vs. the product of molal concentrations (m2m3) of the two amides (Eq. 4). Slopes of linear fits with zero intercept yield chemical potential derivatives quantifying preferential interactions between the two amides.
One-Way Alpha Values for Interactions of Five Amide Solutes with Amide O, N, and C Atoms.
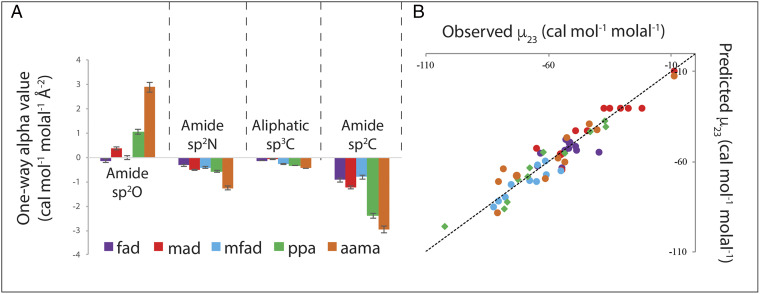
Analysis of the 10 µ23 values determined from Fig. 1 together with previous results for the interactions of these five amides with other amides (31) by Eq. 1 yields one-way alpha values for interactions of these five amides with each of the four types of unified atoms of amide compounds. These one-way alpha values are plotted as bar graphs in Fig. 2 and listed in SI Appendix, Table S2. SI Appendix, Fig. S1 compares one-way alpha value for all 12 amide compounds investigated to date. Fig. 2 and SI Appendix show that all amide compounds investigated here and previously interact favorably with amide sp2C, amide sp2N, and aliphatic sp3C, and that all but urea and formamide interact unfavorably with amide sp2O.
Fig. 2.
(A) Strengths of interaction of five amides (formamide [fad], N-methylformamide [mfad], malonamide [mad], propionamide [ppa] and N-acetylalanine N-methylamide [aama]) with amide and hydrocarbon unified atoms. Bar graphs compare interaction potentials (one-way alpha values; SI Appendix, Table S2) quantifying interactions of these five amide compounds with a unit area of amide sp2O, amide sp2N, aliphatic sp3C, and amide sp2C at 23 °C. Favorable interactions have negative one-way alpha values, while unfavorable interactions have positive one-way alpha values. (B) Comparison of predicted and observed µ23 values for pairwise interactions of these five amide compounds at 23 °C. Predictions of µ23 use one-way alpha values for these five amide compounds with amide sp2O, amide sp2N, amide sp2C, and aliphatic sp3 C from A and SI Appendix, Table S2. Color scheme is that of A. Observed µ23 values are from SI Appendix, Tables S1 and S3. The line represents equality of predicted and observed values.
Strengths of Pairwise Interactions of Amide sp2O, N, C, and Aliphatic sp3C Unified Atoms.
All 105 µ23 values (SI Appendix, Tables S3 and S4) for interactions of 12 different amide compounds with each other, and, in some cases, with naphthalene and/or anthracene, were analyzed using Eq. 3 to obtain 10 two-way alpha values (αsp2O,sp2O, αsp2O,sp2N, αsp2O, sp2C, αsp2O,sp3C; αsp2N,sp2N, αsp2N,sp2C, αsp2N,sp3C; αsp2C,sp2C, αsp2C,sp3C; αsp3C,sp3C). The previous one-way (31) analysis revealed that interactions of sp2C atoms of aromatic hydrocarbons and of amides are similar if not identical, and they are analyzed together here. Alternative analyses of sp2C presented in SI Appendix, section IV justify this treatment. In this analysis of all amide−amide and amide−aromatic µ23 values, the number of equations (105 applications of Eq. 4) exceeds the number of unknowns (10 two-way alpha values) by more than 10-fold, making them highly overdetermined. The hypotheses of additivity and ASA dependence of the various atom−atom contributions to µ23 underlying Eq. 3 are tested quantitatively by comparison of predicted and observed µ23 values (SI Appendix, Tables S3 and S4 and Fig. 3B).
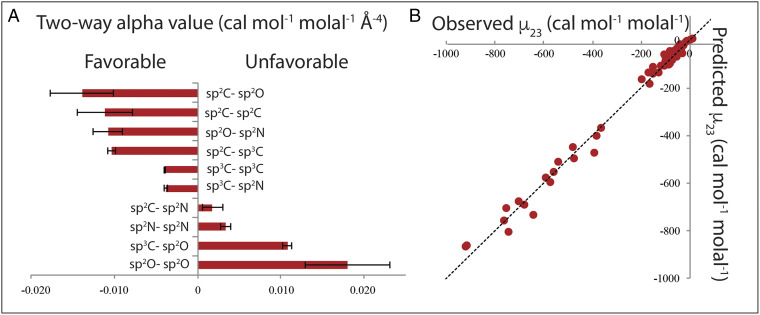
Fig. 3.
Amide compound atom−atom interaction strengths and their ability to predict µ23 values. (A) Bar graph summary of two-way alpha values (Table 1) quantifying pairwise interactions of unified C, N, and O atoms of amide compounds, relative to interactions with water at 23 °C. (Sp2C is combined amide and aromatic sp2C.) Negative two-way alpha values indicate favorable interactions. (B) Comparison of predicted and observed µ23 values for interactions of each pair of amide compounds investigated at 23 °C. Predictions of µ23 use two-way alpha values in Table 1. Observed µ23 values are from SI Appendix, Tables S1 and S3. The line represents equality of predicted and observed values.
Results of this analysis (two-way alpha values) quantifying the pairwise interactions of amide sp2O, sp2N, sp2C, and aliphatic sp3C unified atoms with one another are listed in Table 1. These 10 two-way alpha values are also plotted in the bar graph of Fig. 3A in ranked order from the most negative (most favorable interactions relative to interactions with water) to the most positive (most unfavorable interactions). Uncertainties in these two-way alpha values range from 3 to 30%, except for the small-magnitude interaction of sp2C with sp2N, where the uncertainty is larger (∼70%). These uncertainties do not affect the semiquantitative conclusions of this research.
Table 1.
Ranked intrinsic strengths of atom−atom interactions of amide compounds
| Interaction Type | Amide atom* | Strength | ||
| i | j | Two-way alpha value (cal⋅mol−1⋅molal−1⋅Å−4) | Relative to sp3C–sp3C | |
| Aliphatic sp3C interactions: | sp3C | sp2C | −0.0103 ± 0.0005 | 2.6 |
| sp3C | sp3C | −0.0039 ± 0.0001 | 1.0 | |
| sp3C | sp2N | −0.0038 ± 0.0002 | 1.0 | |
| sp3C | sp2O | 0.0108 ± 0.0005 | (2.8)† | |
| Amide sp2C interactions: | sp2C | sp2O | −0.0139 ± 0.0038 | 3.6 |
| sp2C | sp2C | −0.0111 ± 0.0033 | 2.9 | |
| sp2C | sp3C | −0.0103 ± 0.0005 | 2.6 | |
| sp2C | sp2N | 0.0018 ± 0.0013 | (0.5)† | |
| Amide sp2O interactions: | sp2O | sp2C | −0.0139 ± 0.0038 | 3.6 |
| sp2O | sp2N | −0.0108 ± 0.0017 | 2.8 | |
| sp2O | sp3C | 0.0108 ± 0.0005 | (2.8)† | |
| sp2O | sp2O | 0.0181 ± 0.0051 | (4.6)† | |
| Amide sp2N interactions: | sp2N | sp2O | −0.0108 ± 0.0017 | 2.8 |
| sp2N | sp3C | −0.0038 ± 0.0002 | 1.0 | |
| sp2N | sp2C | 0.0018 ± 0.0013 | (0.5)† | |
| sp2N | sp2N | 0.0034 ± 0.0006 | (0.9)† | |
Labels i and j are interchangeable, so only 10 of the 16 two-way alpha values listed are unique.
Unfavorable interaction strengths (indicated by parentheses) are expressed relative to the magnitude of the sp3C–sp3C interaction.
Six of the 10 atom−atom interactions in Table 1 are favorable, with negative two-way alpha values. The four most favorable interactions, of similar strength when expressed per unit area of each unified atom, are sp2O−sp2C, sp2C−sp2C, sp2O−sp2N, and sp2C−sp3C. Two-way alpha values for these four interactions are about 3 times more negative than two-way alpha values for sp3C−sp3C and sp3C−sp2N interactions. The most unfavorable interaction in this set is sp2O−sp2O. Sp2O−sp3C and sp2N−sp2N interactions are modestly unfavorable, while the sp2N−sp2C interaction is slightly unfavorable. The signs and relative magnitudes of these interaction strengths, relative to interactions with water, make good chemical sense, as discussed below. We therefore conclude that these two-way alpha values not only are useful to predict and interpret interactions of amide solutes and amide effects on protein processes but also have fundamental chemical significance for understanding the weak interactions that drive self-assembly.
Comparison of Observed µ23 Values and One-Way Alpha Values for Amide Solutes with Predictions from Two-Way Alpha Values.
Fig. 3B compares observed µ23 values for interactions of the series of urea and amide solutes with those predicted from two-way alpha values (Table 1) and ASA information (31) using Eq. 3. All these observed and predicted µ23 values are listed in SI Appendix, Tables S3 and S4. For amide−amide interactions, predicted and observed µ23 values are in good agreement within the combined uncertainties ( SD, typically 15%) for about 90% of the interactions investigated (94 out of 105).
SI Appendix, Tables S3 and S5 and Fig. S2 summarizes results of additional comparisons of observed and predicted µ23 values and one-way alpha values for amide solutes investigated here. SI Appendix, Table S3 compares µ23 values for interactions of five amides (formamide, malonamide, N-methylformamide, propionamide, and aama) determined here and previously (31) with predictions of µ23 and µ32 from one-way alpha values using Eq. 1. SI Appendix, Fig. S2 and Table S5 compares one-way alpha values obtained by analysis of sets of µ23 values for amides using Eq. 1 with predictions from two-way alpha values (Table 1) using Eq. 2. In almost all cases, good agreement within the combined uncertainties is obtained.
Applications of Two-Way Alpha Values.
SI Appendix provides applications of two-way alpha values for amide atom−atom interactions to analyze effects of the amide urea and the polyamide polyvinyl pyrrolidone on protein stability and also discusses the potential use of these two-way alpha values in polymer solution thermodynamic analyses of processes like expansion and contraction of polyamide flexible coils and aggregation/droplet formation of intrinsically disordered polypeptides.
Discussion
Chemical Significance of Amide Two-Way Alpha Values.
Interactions of aliphatic sp3C with aliphatic sp3C, amide/aromatic sp2C, and amide sp2N and sp2O.
Interactions of aliphatic sp3C atoms of amide compounds with aliphatic sp3C, amide sp2C, and amide sp2N atoms of other amides are favorable, while interactions with amide sp2O are unfavorable, relative to interactions with water. Strengths (two-way alpha values) of these preferential interactions, expressed per unit area of each atom type (Table 1), span a wide range. These two-way alpha values are well determined from the global fitting, with uncertainties of 3 to 5%.
The preferential interaction of sp3C with sp2C, quantified per unit ASA of each atom type, is one of the four most favorable atom−atom interactions characterized here. This is often called a CH−π interaction, and should also involve a hydrophobic effect from the burial of sp3C and sp2C ASA when it occurs. From Table 1 two-way alpha values, the strength of a favorable sp3C−sp2C interaction is almost 3 times that of an sp3C−sp3C interaction, which is presumably driven by a hydrophobic effect from removing sp3C ASA from water. Interpreted most simply, this comparison indicates that the CH−π component of the favorable interaction of aliphatic sp3C with amide or aromatic sp2C contributes more than the hydrophobic component of this interaction.
Two-way alpha values in Table 1 also reveal that the sp3C−sp2N preferential interaction is about as favorable as the sp3C−sp3C interaction. Because sp2N unified atoms are expected to interact more favorably with water than sp3C atoms do, it follows that the intrinsic interaction of sp3C with sp2N (i.e., not relative to water) is more favorable than the intrinsic interaction of sp3C with sp3C.
The sp3C−sp2O interaction in water is highly unfavorable, with a two-way alpha value that is equal in magnitude and opposite in sign to the sp3C−sp2C interaction. An unfavorable interaction means that intrinsic interactions of the unified sp3C and sp2O atoms with water are more favorable than the intrinsic sp3C−sp2O interaction. The sp3C−sp2O interaction is unfavorable because the intrinsic interaction of water with sp2O is favorable while the intrinsic interaction of sp3C with amide sp2O is probably comparably unfavorable to its intrinsic interaction with water.
Amide/aromatic sp2C interactions.
From Table 1, interactions of amide/aromatic sp2C atoms with amide sp2O, aliphatic sp2C, and amide sp3C atoms of other amides are all very favorable, while interactions with amide sp2N are slightly unfavorable, relative to interactions with water. Overall, amide/aromatic sp2C atoms interact more favorably with the atoms of amide compounds than any other atom type in Table 1. These sp2C two-way alpha values are not as accurately known as sp3C two-way alpha values. Except for the sp2C−sp3C interaction (5% uncertainty), uncertainties are 27% for interactions with sp2O and sp2C and 70% for the very weak interaction with sp2N.
The sp2C−sp2O interaction is the most favorable interaction quantified here, with a two-way alpha value which is about 3 times as favorable as for hydrophobic sp3C−sp3C, which we take as a reference. In all likelihood, the sp2C−sp2O interaction is an n–π* interaction [one example of an lp–π interaction (11)] involving n-shell electrons of amide sp2O and the π-system of the amide group or aromatic ring, as characterized previously in structural and spectroscopic studies and molecular dynamics simulations (12–17). The observation that a single two-way alpha value quantifies this interaction for both amide and aromatic sp2C is a compelling argument for the use of ASA in this analysis. This two-way alpha value is very similar to that deduced from the one-way alpha value for the interaction of naphthalene with amide sp2O (31) (SI Appendix, Table S6). Water forms hydrogen bonds to amide sp2O atoms and presumably participates in an lp–π interaction with sp2C atoms, so the strength of the sp2O−sp2C interaction in Table 1 is relative to these competitive interactions involving water.
Comparison of two-way alpha values in Table 1 reveals that the sp2C−sp2C interaction is about as favorable, per unit area of each participant, as the sp2C−sp3C interaction discussed above. Therefore it is likely that the π–π component of the sp2C−sp2C interaction, expressed per unit area of each participant, is similar in strength to the CH−π interaction and contributes about twice as much as the hydrophobic effect to the favorable sp2C−sp2C interaction.
The unnamed interaction of amide/aromatic sp2C with amide sp2N is very marginally unfavorable. This interaction is not as favorable as the sp3C−sp2N interaction, probably because the intrinsic interaction of water oxygen lps with the sp2C π-system is more favorable than the interaction of water with sp3C. Even so, because it is only marginally unfavorable, there should be no significant free energy penalty for forming contacts between amide/aromatic sp2C and amide sp2N in a protein interface.
Amide sp2O interactions.
From Table 1, interactions of amide sp2O atoms with amide and aromatic sp2C and amide sp2N atoms are both very favorable, while amide sp2O interactions with aliphatic sp3C and amide sp2O are very unfavorable, relative to interactions with water. Uncertainties in these two-way alpha values are moderate, ranging from 5% for sp2O−sp3C to 28% for sp2O−sp2C and sp2O−sp2O.
The favorable interaction of amide sp2O with amide/aromatic sp2C is discussed above. The similarly favorable interaction of amide sp2O with amide sp2N in water is almost certainly the NH···O=C hydrogen bond interaction in which the unified amide sp2N atom is the donor and the sp2O atom is the acceptor (3–6). The amide sp2O−amide sp2O interaction is almost twice as unfavorable as the amide sp2O–aliphatic sp3C interaction, because of the very favorable intrinsic interaction of amide sp2O with water, which contributes twice as much in magnitude to the two-way alpha value for sp2O−sp2O as for sp2O−sp3C.
Amide sp2N interactions.
From Table 1, the amide sp2N−amide sp2O interaction is the most favorable interaction involving amide sp2N, while the amide sp2N−aliphatic sp3C interaction is modestly favorable, and the amide sp2N interactions with amide/aromatic sp2C and amide sp2N is slightly unfavorable, relative to interactions with water. Uncertainties in two-way alpha values for interactions involving amide sp2N are small (5 to 18%) except for the very weak interaction with sp2C. All these interactions except amide sp2N−amide sp2N are discussed above.
The amide sp2N−amide sp2N interaction, which very likely is the NH···N hydrogen bond, is modestly unfavorable, indicating that hydrogen bonding of amide sp2N with water is intrinsically more favorable. Consistent with this, NH···N hydrogen bonds are seldom observed in protein secondary structures, except involving proline (48). However, a hydrogen bond between unified N atoms of heterocyclic aromatic rings occurs in both AT (also AU) and GC base pairs of nucleic acid duplexes.
Conclusion
Average strengths of interaction of amide O, N, and C unified atoms, quantified per unit of accessible area of each atom by two-way alpha values, provide important bridges between protein structural (ASA) information, molecular dynamics simulations, and experimental studies of protein−solute interactions and solute effects of protein processes, as well as a window into a chemistry of weak interactions of these O, N, and C unified atoms in water.
Materials and Methods
Details about materials, vapor pressure osmometry (VPO) measurements, and data analysis (determination of μ23, surface area calculations) are provided in SI Appendix, section V and ref. 31.
Supplementary Material
Acknowledgments
We thank Prof. Gary Pielak, Emily Zytkiewicz, and the reviewers for their comments on the manuscript, and gratefully acknowledge support of NIH Grant GM R35-118100 for this research.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012481117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Timasheff S. N., . ““Control of protein stability and reactions by weakly interacting cosolvents: The simplicity of the complicated”” in Advances in Protein Chemistry: Linkage Thermodynamics of Macromolecular Interactions, DiCera E., Ed. (Elsevier Academic, San Diego, CA, 1998), Vol. vol. 51, pp. 355–432. [DOI] [PubMed] [Google Scholar]
- 2.Record M. T. Jr, Guinn E., Pegram L., Capp M., Introductory lecture: Interpreting and predicting Hofmeister salt ion and solute effects on biopolymer and model processes using the solute partitioning model. Faraday Discuss. 160, 9–44 (2013), and discussion 160, 103–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dill K. A., Dominant forces in protein folding. Biochemistry 29, 7133–7155 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Kortemme T., Morozov A. V., Baker D., An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J. Mol. Biol. 326, 1239–1259 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Nick Pace C., Scholtz J. M., Grimsley G. R., Forces stabilizing proteins. FEBS Lett. 588, 2177–2184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newberry R. W., Raines R. T., A prevalent intraresidue hydrogen bond stabilizes proteins. Nat. Chem. Biol. 12, 1084–1088 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanford C., The hydrophobic effect and the organization of living matter. Science 200, 1012–1018 (1978). [DOI] [PubMed] [Google Scholar]
- 8.Spolar R. S., Record M. T. Jr., Coupling of local folding to site-specific binding of proteins to DNA. Science 263, 777–784 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Baldwin R. L., Properties of hydrophobic free energy found by gas-liquid transfer. Proc. Natl. Acad. Sci. U.S.A. 110, 1670–1673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldwin R. L., Dynamic hydration shell restores Kauzmann’s 1959 explanation of how the hydrophobic factor drives protein folding. Proc. Natl. Acad. Sci. U.S.A. 111, 13052–13056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novotný J., Bazzi S., Marek R., Kozelka J., Lone-pair-π interactions: Analysis of the physical origin and biological implications. Phys. Chem. Chem. Phys. 18, 19472–19481 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Bartlett G. J., Choudhary A., Raines R. T., Woolfson D. N., n–>π* interactions in proteins. Nat. Chem. Biol. 6, 615–620 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartlett G. J., Newberry R. W., VanVeller B., Raines R. T., Woolfson D. N., Interplay of hydrogen bonds and n→π* interactions in proteins. J. Am. Chem. Soc. 135, 18682–18688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newberry R. W., VanVeller B., Guzei I. A., Raines R. T., n→π* interactions of amides and thioamides: Implications for protein stability. J. Am. Chem. Soc. 135, 7843–7846 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh S. K., Das A., The n → π* interaction: A rapidly emerging non-covalent interaction. Phys. Chem. Chem. Phys. 17, 9596–9612 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Newberry R. W., Raines R. T., The n→π* interaction. Acc. Chem. Res. 50, 1838–1846 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newberry R. W., Raines R. T., Secondary forces in protein folding. ACS Chem. Biol. 14, 1677–1686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer E. A., Castellano R. K., Diederich F., Interactions with aromatic rings in chemical and biological recognition. Angew. Chem. Int. Ed. Engl. 42, 1210–1250 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Neel A. J., Hilton M. J., Sigman M. S., Toste F. D., Exploiting non-covalent π interactions for catalyst design. Nature 543, 637–646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Courtenay E. S., Capp M. W., Anderson C. F., Record M. T. Jr., Vapor pressure osmometry studies of osmolyte-protein interactions: Implications for the action of osmoprotectants in vivo and for the interpretation of “osmotic stress” experiments in vitro. Biochemistry 39, 4455–4471 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Plevin M. J., Bryce D. L., Boisbouvier J., Direct detection of CH/π interactions in proteins. Nat. Chem. 2, 466–471 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Nishio M., Umezawa Y., Fantini J., Weiss M. S., Chakrabarti P., CH-π hydrogen bonds in biological macromolecules. Phys. Chem. Chem. Phys. 16, 12648–12683 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Hudson K. L.et al., Carbohydrate-aromatic interactions in proteins. J. Am. Chem. Soc. 137, 15152–15160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pegram L. M., Record M. T. Jr., Thermodynamic origin of Hofmeister ion effects. J. Phys. Chem. B 112, 9428–9436 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capp M. W.et al., Interactions of the osmolyte glycine betaine with molecular surfaces in water: Thermodynamics, structural interpretation, and prediction of m-values. Biochemistry 48, 10372–10379 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guinn E. J., Pegram L. M., Capp M. W., Pollock M. N., Record M. T. Jr., Quantifying why urea is a protein denaturant, whereas glycine betaine is a protein stabilizer. Proc. Natl. Acad. Sci. U.S.A. 108, 16932–16937 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diehl R. C., Guinn E. J., Capp M. W., Tsodikov O. V., Record M. T. Jr., Quantifying additive interactions of the osmolyte proline with individual functional groups of proteins: Comparisons with urea and glycine betaine, interpretation of m-values. Biochemistry 52, 5997–6010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guinn E. J.et al., Quantifying functional group interactions that determine urea effects on nucleic acid helix formation. J. Am. Chem. Soc. 135, 5828–5838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knowles D. B.et al., Chemical interactions of polyethylene glycols (PEGs) and glycerol with protein functional groups: Applications to effects of PEG and glycerol on protein processes. Biochemistry 54, 3528–3542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng X.et al., Basis of protein stabilization by K glutamate: Unfavorable interactions with carbon, oxygen groups. Biophys. J. 111, 1854–1865 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng X.et al., Experimental atom-by-atom dissection of amide-amide and amide-hydrocarbon interactions in H2O. J. Am. Chem. Soc. 139, 9885–9894 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng X.et al., Quantifying interactions of nucleobase atoms with model compounds for the peptide backbone and glutamine and asparagine side chains in water. Biochemistry 57, 2227–2237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Naim A., Inversion of Kirkwood-Buff theory of solutions—Application to water-ethanol system. J. Chem. Phys. 67, 4884–4890 (1977). [Google Scholar]
- 34.Chitra R., Smith P. E., Preferential interactions of cosolvents with hydrophobic solutes. J. Phys. Chem. B 105, 11513–11522 (2001). [Google Scholar]
- 35.Smith P. E., Chemical potential derivatives and preferential interaction parameters in biological systems from Kirkwood-Buff theory. Biophys. J. 91, 849–856 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canchi D. R., Garcia A. E., . ““Cosolvent effects on protein stability”” in Annual Review of Physical Chemistry, Johnson M. A., Martinez T. J., Eds. (Annual Reviews, Palo Alto, CA, 2013), Vol. vol. 64, pp. 273–293. [DOI] [PubMed] [Google Scholar]
- 37.Trzesniak D., van der Vegt N. F. A., van Gunsteren W. F., Computer simulation studies on the solvation of aliphatic hydrocarbons in 6.9 M aqueous urea solution. Phys. Chem. Chem. Phys. 6, 697–702 (2004). [Google Scholar]
- 38.Trzesniak D., Van Der Vegt N. F. A., Van Gunsteren W. F., Analysis of neo-pentane-urea pair potentials of mean force in aqueous urea. Mol. Phys. 105, 33–39 (2007). [Google Scholar]
- 39.Ma L., Pegram L., Record M. T. Jr., Cui Q., Preferential interactions between small solutes and the protein backbone: A computational analysis. Biochemistry 49, 1954–1962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganguly P., van der Vegt N. F. A., Representability and transferability of kirkwood-buff iterative Boltzmann inversion models for multicomponent aqueous systems. J. Chem. Theory Comput. 9, 5247–5256 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Lin B., Lopes P. E. M., Roux B., MacKerell A. D. Jr., Kirkwood-Buff analysis of aqueous N-methylacetamide and acetamide solutions modeled by the CHARMM additive and Drude polarizable force fields. J. Chem. Phys. 139, 084509 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nozaki Y., Tanford C., The solubility of amino acids and related compounds in aqueous urea solutions. J. Biol. Chem. 238, 4074–4081 (1963). [PubMed] [Google Scholar]
- 43.Auton M., Holthauzen L. M. F., Bolen D. W., Anatomy of energetic changes accompanying urea-induced protein denaturation. Proc. Natl. Acad. Sci. U.S.A. 104, 15317–15322 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auton M., Rösgen J., Sinev M., Holthauzen L. M. F., Bolen D. W., Osmolyte effects on protein stability and solubility: A balancing act between backbone and side-chains. Biophys. Chem. 159, 90–99 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guinn E. J., Kontur W. S., Tsodikov O. V., Shkel I., Record M. T. Jr., Probing the protein-folding mechanism using denaturant and temperature effects on rate constants. Proc. Natl. Acad. Sci. U.S.A. 110, 16784–16789 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Courtenay E. S., Capp M. W., Saecker R. M., Record M. T. Jr., Thermodynamic analysis of interactions between denaturants and protein surface exposed on unfolding: Interpretation of urea and guanidinium chloride m-values and their correlation with changes in accessible surface area (ASA) using preferential interaction coefficients and the local-bulk domain model. Proteins 41, 72–85 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Pegram L. M., Record M. T. Jr., Hofmeister salt effects on surface tension arise from partitioning of anions and cations between bulk water and the air-water interface. J. Phys. Chem. B 111, 5411–5417 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Deepak R. N. V. K., Sankararamakrishnan R., Unconventional N-H…N hydrogen bonds involving proline backbone nitrogen in protein structures. Biophys. J. 110, 1967–1979 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.