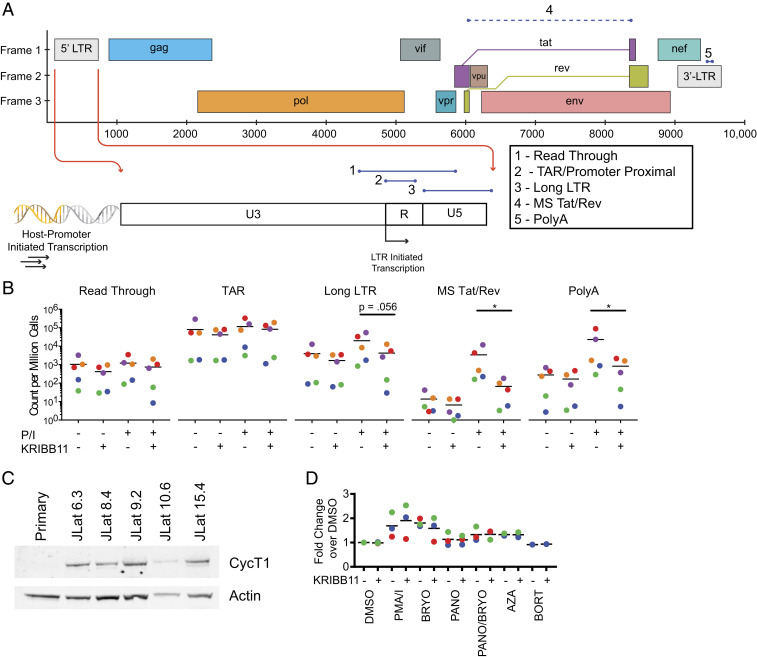
Fig. 4.
Analysis of transcriptional elongation in HIV-1 latency reversal. (A) Evaluation of steps in HIV-1 transcription using a panel of RT-ddPCR assays (55) that target five different regions of HIV-1 transcripts. The read-through assay includes HIV-1 sequences upstream of the HIV-1 transcription start site (TSS) and quantifies transcripts resulting from host-promoter–initiated transcription. The TAR assay targets the TAR region directly downstream from the HIV-1 TSS. The Long LTR assay starts downstream of the HIV-1 TSS and ends just past the 5′ LTR. The MS Tat/Rev and PolyA assays target distal sites in HIV-1 transcripts and require correct splicing and termination, respectively. (B) Analysis of HIV-1 transcript levels in resting CD4+ T cells from treated patients with or without PMA/I stimulation in the presence or absence of KRIBB11. Counts of various transcript types are quantified and normalized per million cells as determined by TERT cell equivalents as described (55). (C) Western blot analysis for CycT1 in whole-cell lysates of resting CD4+ T cells and clones of J-Lat cells. (D) Intracellular flow cytometry analysis of CycT1 levels in resting CD4+ T cells from treated patients after stimulation with candidate LRAs. Median fluorescence intensity is plotted as a fold change over DMSO control.