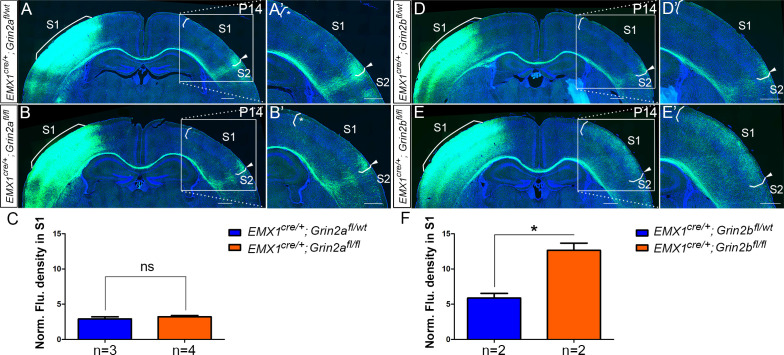
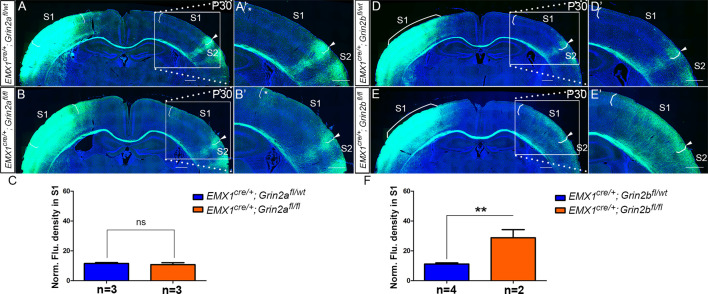
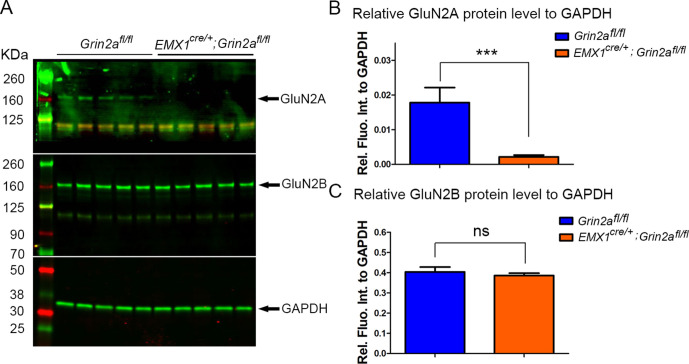
Figure 6. Emx1cre/+; Grin2bfl/fl but not Emx1cre/+; Grin2afl/fl mice had the same disrupted callosal innervation patterns as Emx1cre/+; Grin1fl/fl at P14.
Callosal innervation patterns in control Emx1cre/+; Grin2afl/wt (A) and Emx1cre/+; Grin2afl/fl mice (B) at P14. ‘*’ points out M1/S1 border. (C) Quantification of fluorescence density. p=0.392. Callosal innervation patterns in control Emx1cre/+; Grin2bfl/wt (D) and Emx1cre/+; Grin2bfl/fl mice (E) at P14. (F) Quantification of fluorescence density. p=0.03. Scale bar: 500 μm for all images.