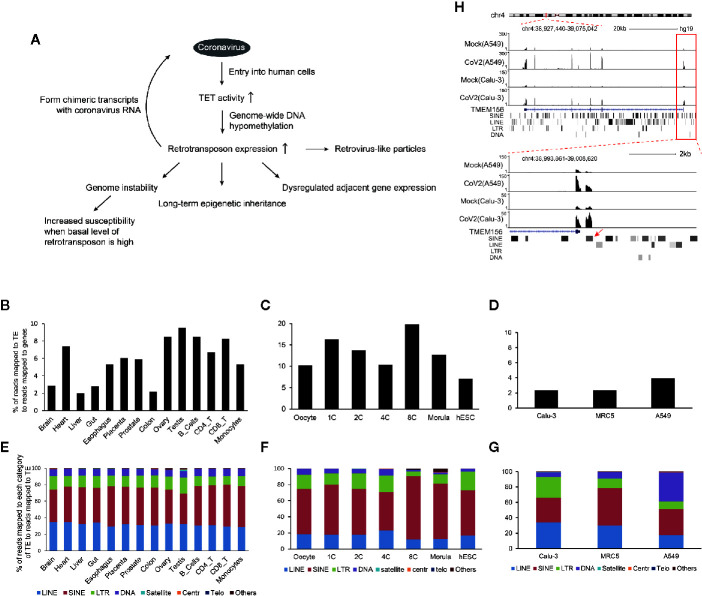
Figure 5.
Model of how coronavirus may impact retrotransposons to harm human cells. (A) Generally, entry of coronavirus into human cells enhances ten-eleven translocation (TET) activity for genome-wide DNA hypomethylation to facilitate retrotransposon enhancement. This may lead to increased fusion between viral and retrotransposon transcripts, reduced genome stability, and increased susceptibility of aged people and cancer patients, dysregulated TE-adjacent gene expression, and this influence may be inherited for a long term. Meanwhile, increased retrovirus-like particles of retrotransposons may be induced. (B–G) Endogenous retrotransposon expression and distribution of subfamilies are variable in human tissues and cells. Bar graphs indicate percentages of reads mapped to TE to reads mapped to genes in human tissues and immunocytes (B), human oocytes and early embryos (C), and normal Calu-3, MRC5 and A549 cells (D). Bar charts (E–G) demonstrate distribution of individual subfamilies of TE in tissues or cell types shown above. (H) UCSC genome browser view of an example of retrotransposon-initiated gene expression by readthrough mechanism.