Abstract
Exosomal microRNA (miRNA) secretion has been characterized as a vital factor in intercellular communication among cancer cells. However, little is known about cancer-secreted miRNAs specifically involved in metastasis of colorectal cancer (CRC). Here, we found that exosomes derived from metastatic CRC cell line SW620 promoted migration, invasion, and epithelial-mesenchymal transition (EMT) of CRC cells. The profiling of exosome miRNAs revealed that microRNA (miR)-335-5p was highly expressed in exosomes from metastatic SW620 cells compared to those derived from primary SW480 cells. miR-335-5p was transmitted from metastatic SW620 cells to CRC cells via exosomes and promoted migration, invasion, and EMT of CRC cells. Moreover, exosome-transmitted miRNA-335-5p promotes CRC cell invasion and metastasis by facilitating EMT via targeting RAS p21 protein activator 1 (RASA1). Overexpression of RASA1 abolished the promotive effects of exosomal miR-335-5p on CRC cell migration, invasion, and EMT. Collectively, our data revealed that exosomal miR-335-5p derived from metastatic CRC cells promotes CRC cell invasion and metastasis by facilitating EMT via targeting RASA1, which may serve as a potential therapeutic target for CRC metastasis.
Keywords: colorectal cancer, exosomes, miR-335-5p, RASA1, metastasis
Graphical Abstract
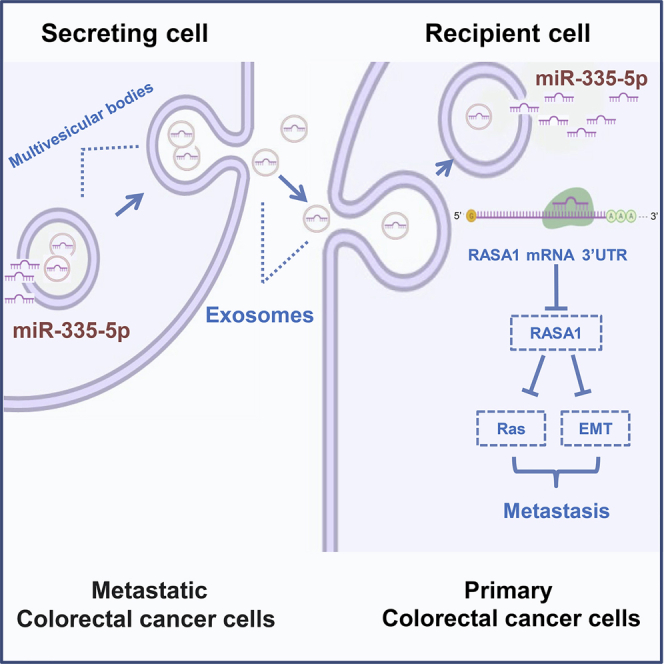
In this study, these data revealed that exosomal miR-335-5p derived from metastatic CRC cells promoted CRC cell invasion and metastasis by facilitating EMT via targeting RASA1, which may serve as a potential therapeutic target for CRC metastasis.
Introduction
Colorectal cancer (CRC) is prevalent throughout the world, with mortality mostly caused by its metastasis to distant critical organs.1,2 Metastasis of CRC is correlated with poor prognosis and indicates terminal stages of cancer.3,4 Approximately one-third of CRC patients will finally develop metastatic disease and are at high risk of recurrence after surgery.5,6 Thus, a better understanding of the exact molecular mechanisms underlying CRC metastasis is important to identify novel diagnostic biomarkers and develop therapeutic strategies for CRC patients.
Recently, exosomes have emerged as an appreciated fundamental aspect of cancer metastasis research. These are endosomes distinguished by size, ranging from 30 to 100 nm, that are secreted from the membrane as multivesicular bodies and contain proteins, lipids, DNA, mRNA, and microRNAs (miRNAs).7 Interestingly, the major part of exosomal RNA is constituted of miRNAs, which serve as genetic messengers and play important roles in regulating gene expression of target cells.8 Exosome secretion has been documented in cancer cells and has been pronounced responsible for priming the formation of a pre-metastatic microenvironment that further promotes engraftment and survival of cancer.9,10 Melo et al.11 and Kobayashi et al.12 reported that serum exosomes are reliable markers for early diagnosis of cancer.
miRNAs are small (about 22 nucleotides) non-coding and single-stranded RNA molecules found in animals or plants and function in the post-transcriptional regulation of gene expression.13,14 Functionally, miRNAs may suppress or promote various kinds of cancers, depending on whether the miRNAs specifically target potential tumor suppressors or oncogenes.15 Recent studies have revealed miRNAs as key actors in cancer metastasis.16,17 What’s more, some researchers have highlighted that miRNAs may have a role in regulating epithelial-to-mesenchymal transition (EMT),18,19 a morphogenetic process by which cells lose their epithelial properties and gain mesenchymal characteristics.
Exosomes not only transfer membrane components between cells but also transfer nucleic acids,20 with miRNAs being a major RNA component of exosomes.21 Exosomal miRNAs secreted by cancer cells can be transferred to recipient cells to regulate gene expression.8,22,23 Recent studies showed that cancer-secreted miRNAs may play an important role in regulating various cellular components of the tumor microenvironment to promote metastasis.8,24 However, little is known about the involvement of cancer-secreted miRNAs in metastasis.
In this study, we found that exosomes derived from metastatic CRC SW620 cells promoted CRC cell migration, invasion, and EMT when compared to exosomes derived from primary SW480 cells. Expression profiling of exosome miRNAs revealed that microRNA (miR)-335-5p was more highly expressed in exosomes from metastatic SW620 cells than in those from SW480 cells. Moreover, we revealed exosome shuttling of miR-335-5p as an underlying regulatory mechanism that promotes CRC cell migration, invasion, and EMT through targeting RAS p21 protein activator 1 (RASA1), a candidate suppressor of RAS function. This discovery could provide a significant basis for the development of novel therapeutic strategies in CRC patients.
Results
Characterization of exosomes derived from CRC cells
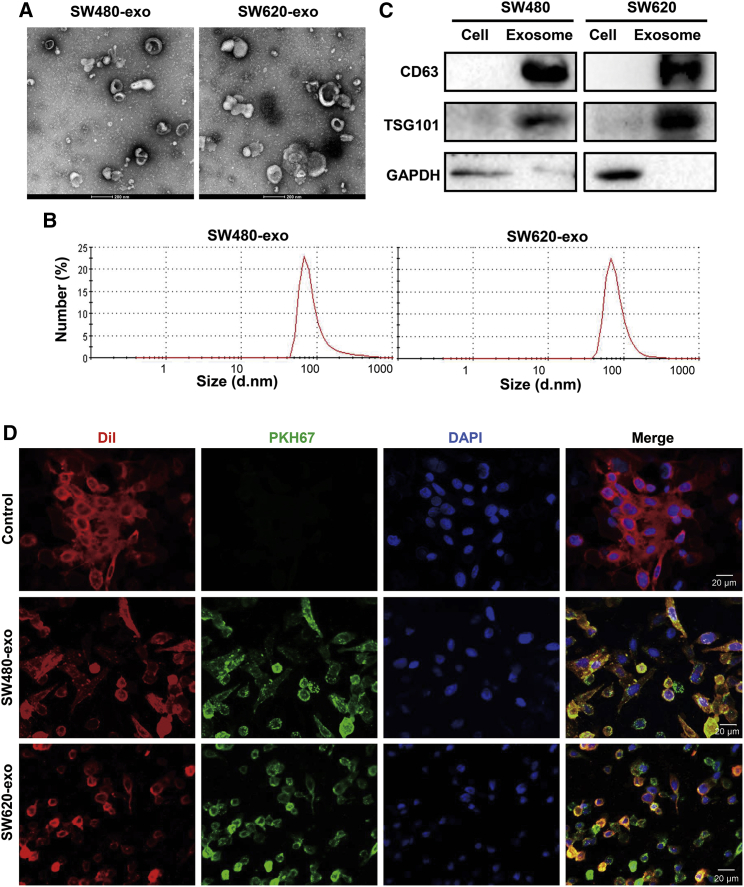
CRC cells shed from their surface a variety of vesicles, some of which are exosomes (size smaller than 100 nm). We performed electron microscopy, nanoparticle size potential analysis, and western blot analysis to characterize vesicles recovered from CRC conditioned medium by differential ultracentrifugation (110,000 × g for 2 h). As expected, we observed 30−100 nm vesicles that were morphologically consistent with exosomes (Figures 1A and 1B). Protein extraction from exosomes, followed by western blotting, demonstrated that these vesicles expressed the exosomal markers CD63 and TSG101 (Figure 1C). To determine whether exosomes can be internalized by CRC cells, we labeled exosomes with the green lipophilic fluorescent dye PKH67 and subsequently co-cultured them with SW480 cells. We found that SW480 cells exhibited high uptake efficiency, as indicated by a fluorescent green signal (Figure 1D). After 24 h incubation with PKH67-labeled exosomes, more than 90% of SW480 cells were positive for PKH67 fluorescence, suggesting that the exosomes were internalized by SW480 cells.
Figure 1.
Characterization of exosomes derived from colorectal cancer (CRC) cell lines
(A) Transmission electron microscopy of exosomes isolated from CRC cell lines SW480 and SW620. Scale bars, 200 nm. (B) Size distribution of isolated exosomes. (C) Western blot analysis of the exosome marker proteins CD63 and TSG101 in SW480 and SW620 cells and their exosomes. (D) Presence of PKH67 lipid dye in SW480 cells treated with PKH67-labeled exosomes. Exosomes were derived from SW480 or SW620 (SW480-exo or SW620-exo) cells and incubated with SW480 cells for 24 h. SW480 cells without treatment were used as the control. Scale bars, 20 μm.
Metastatic CRC SW620 cell-secreted exosomes promote migration, invasion, and EMT of CRC cells
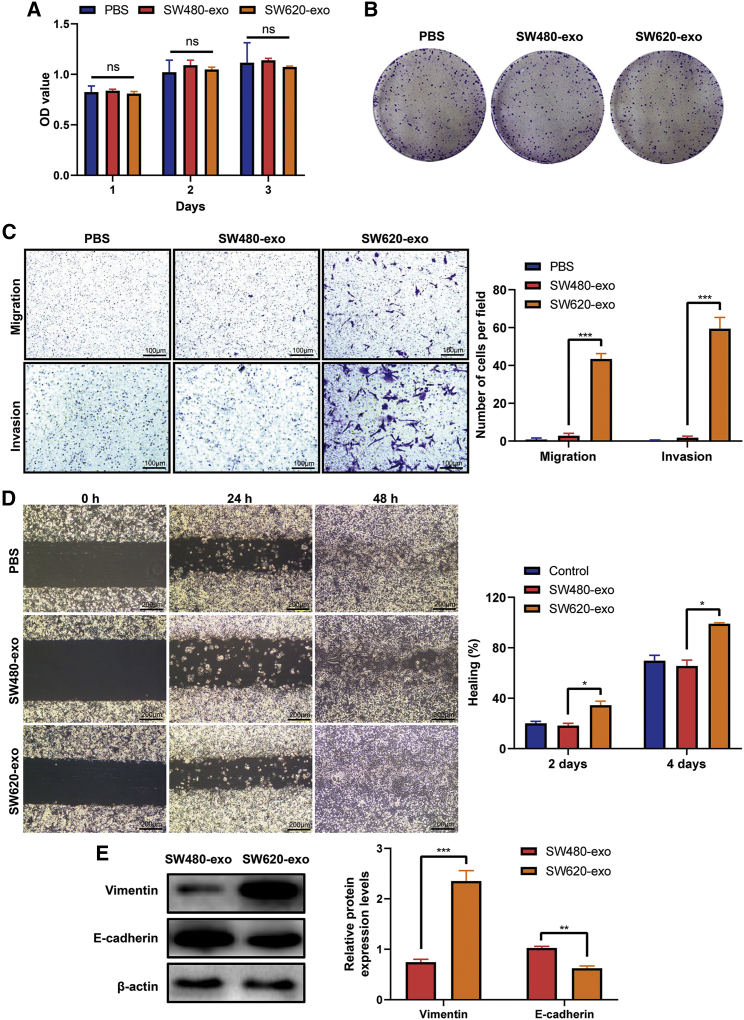
We used as experimental models the cell lines SW480 and SW620, which are derived from primary (SW480) and metastatic (SW620) lesions from a single colon cancer patient.13 SW480 cells were incubated with exosomes derived from SW480 or SW620 (SW480-exo or SW620-exo) cells, and cell viability was determined at 24 h, 48 h, and 72 h with Cell Counting Kit 8 (CCK-8). The results showed that SW480-exo or SW620-exo cells had no significant effects on cell growth (Figure 2A). Also, plate colony-formation assays revealed no difference in growth between SW480-exo or SW620-exo cells (Figure 2B). However, exosomes derived from metastatic CRC SW620 cells significantly promoted the migration and invasion of SW480 cells through Transwell assays (Figures 2C and S1). In addition, SW620-exo improved the wound-healing ability of SW480 cells when compared with SW480-exo and control (phosphate-buffered saline [PBS]) groups (Figure 2D). Next, we performed western blot analysis to determine expression levels of proteins associated with EMT. The results illustrated that SW620-exo treatment significantly increased vimentin, fibronectin, and N-cadherin expression and decreased E-cadherin expression relative to SW480-exo treatment (Figures 2E and S2), indicating that exosomes derived from metastatic CRC cells induced EMT in SW480 cells.
Figure 2.
Exosomes derived from the metastatic CRC cell line SW620 promote CRC cell migration, invasion, and epithelial-mesenchymal transition (EMT)
(A) Viability of SW480 cells treated with PBS, SW480-exo, or SW620-exo for the indicated time; viability was assessed using a Cell Counting Kit-8 assay. ns, not significant. Data are presented as mean ± SD (n = 3). (B) Colony-formation abilities of SW480 cells treated with PBS, SW480-exo, or SW620-exo, as detected by a colony-forming assay. (C) Migration and invasion abilities of SW480 cells treated with PBS, SW480-exo, or SW620-exo, as detected using a Transwell assay. Representative images from triplicate experiments are shown. Data are presented as mean ± SD (n = 3). ∗∗∗p < 0.001. Scale bars, 100 μm. (D) Wound-healing assays of SW480 cells treated with PBS, SW480-exo, or SW620-exo. Data are presented as mean ± SD (n = 3). ∗p < 0.05. Scale bars, 200 μm. (E) Western blot analysis of vimentin and E-cadherin protein expression levels in SW480 cells treated with SW480-exo or SW620-exo. Data are presented as mean ± SD (n = 3). ∗∗p < 0.01, ∗∗∗p < 0.001.
miR-335-5p is highly expressed in exosomes derived from metastatic CRC cells
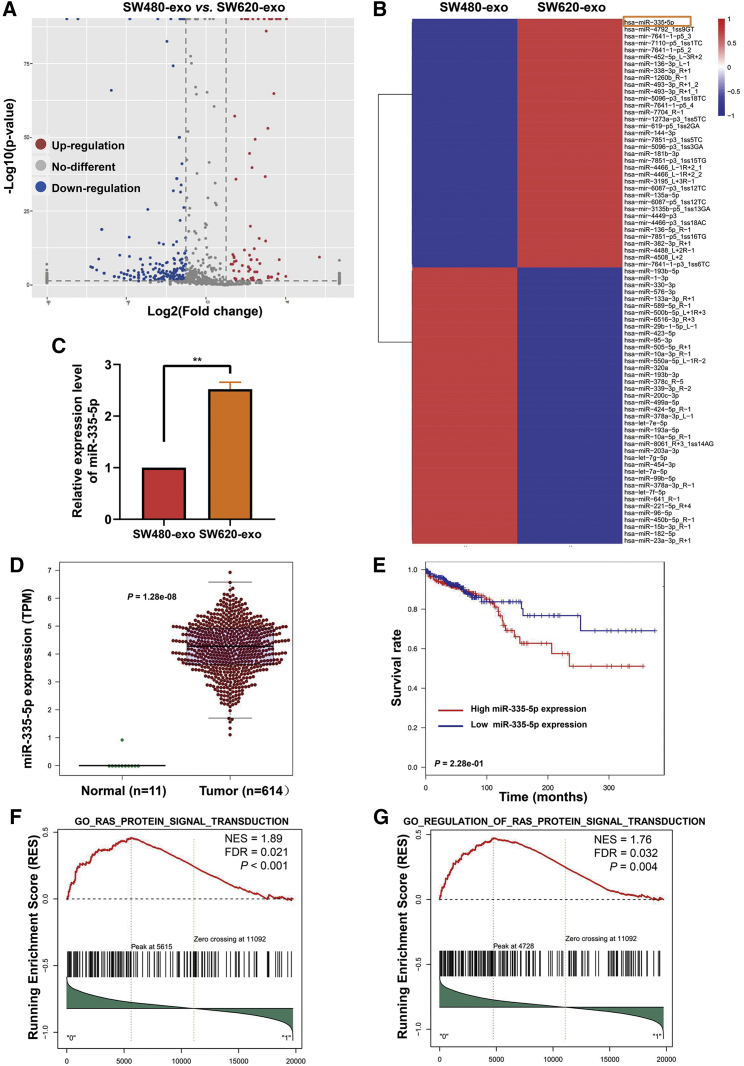
miRNA sequencing was performed to determine miRNA expression profiles in SW480-exo or SW620-exo. We found SW480-exo and SW620-exo to have significantly different miRNA expression profiles (Figure 3A) and identified 77 miRNAs as having a fold change greater than 5 (Figure 3B). Through quantitative PCR, we verified that miR-335-5p was more highly expressed in SW620-exo than in SW480-exo (Figure 3C). We also have profiled the expression level of miR-335-5p in six CRC cell lines (SW620, DLD1, SW480, HCT116, HT29, and Caco-2) through quantitative PCR and showed the expression level of miR-335-5p was higher in the SW620 cell line than the SW480 cell line (Figure S3). With the use of miRNA and mRNA expression data from The Cancer Genome Atlas (TCGA), we confirmed that miR-335-5p was significantly elevated in CRC relative to normal samples (Figure 3D), and cases with high expression of miR-335-5p had a poorer overall survival rate (Figure 3E). We also analyzed TCGA data and found that the expression level of miR-335-5p was significantly elevated in metastatic rectal cancer compared with non-metastatic patients (p = 0.0297; Figure S4A). There was also a slightly elevated in metastatic colon cancer compared with non-metastatic patients, although there was no significance difference (p = 0.137; Figure S4B). In addition, to explore the function of miR-335-5p in CRC, we performed gene set enrichment analysis (GSEA) analysis on the genes expressed in CRC samples having high or low expression of miR-335-5p. For each gene set, we tested whether the genes were enriched in any direction and set the number of rotations to 10,000. We identified significant enrichment in two biological pathways: Ras protein signal transduction and regulation of Ras protein signal transduction (false discovery rate [FDR] < 0.05) (Figures 3F and 3G). These results indicate that SW620 exosomes have higher levels of miR-335-5p, and the functions of miR-335-5p in CRC may relate to activation of the Ras signaling pathway.
Figure 3.
Expression of miR-335-5p is significantly higher in SW620-exo than in SW80-exo
(A) Volcano plot of differentially expressed miRNAs in SW480-exo and SW620-exo. (B) Heatmap of differentially expressed miRNAs in SW480-exo and SW620-exo (fold change ≥ 5.0). (C) Quantitative RT-PCR analysis of miR-335-5p expression in SW480-exo and SW620-exo. Data are presented as mean ± SD (n = 3). ∗∗p < 0.01. (D) Expression of miR-335-5p in normal and CRC samples based on analysis of The Cancer Genome Atlas (TCGA) data. Data are presented as mean ± SD. p = 1.28e-08. (E) Survival analysis of CRC patients expressing high and low levels of miR-335-5p. p = 2.28e-01 (F) Gene-set enrichment analysis (GSEA) analysis showed that the differentially expressed genes associated with miR-335-5p level in CRC samples were enriched for the Ras protein signal transduction pathway. p < 0.001. (G) GSEA analysis showed that the differentially expressed genes associated with miR-335-5p level in CRC samples were enriched for the regulation of Ras protein signal transduction pathway. p = 0.004
miR-335-5p promotes migration and invasion of CRC cells by targeting RASA1
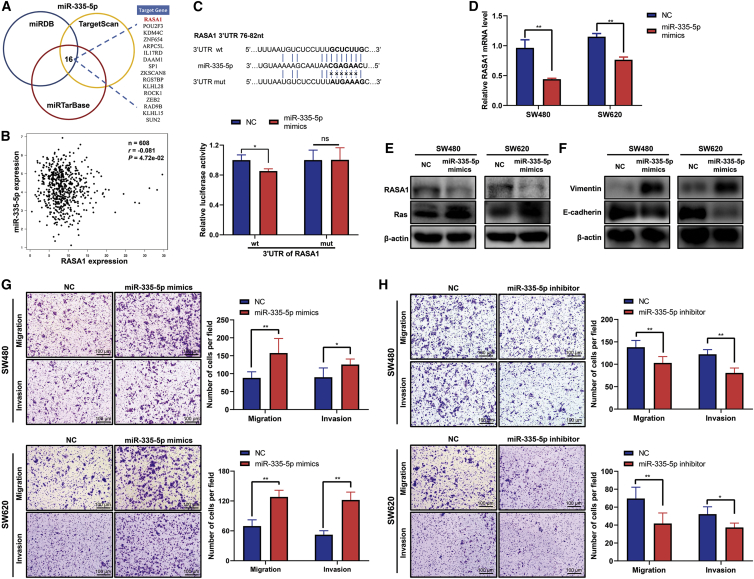
The potential target genes of miR-335-5p were predicted by three bioinformatics algorithms: TargetScan, miRDB, and miRTarBase (Figure 4A). Interestingly, RASA1, a negative regulator of the Ras signaling pathway, is one of the potential targets of miR-335-5p. Analysis of data downloaded from TCGA indicated that expression of RASA1 was significantly associated with miR-335-5p level in CRC patients (r = −0.081, p = 0.00472) (Figure 4B). Figure 4C shows a putative miR-335-5p target site in the 3′ UTR of RASA1 identified by bioinformatic analysis. Luciferase reporter experiments in SW480 cells using the wild-type (WT) RASA1 3′ UTR showed a significant decrease in luciferase activity upon transfection of miR-335-5p mimics (Figure 4C). In contrast, the luciferase activity of a reporter incorporating the mutant (mut) RASA1 sequence was not affected by miR-335-5p (Figure 4C), suggesting that miR-335-5p directly targets that specific region of the 3′ UTR of RASA1.
Figure 4.
miR-335-5p inhibits CRC cell migration, invasion, and EMT by targeting RASA1
(A) Prediction of target genes of miR-335-5p using three computational target prediction algorithms (TargetScan 50, miRDB, and miRTarBase). (B) Correlation analysis of miR-335-5p and RASA1 mRNA expression levels in CRC tissues based on TCGA data. ∗p < 0.05. (C) Wild-type (WT) and mutant (mut) binding sites of miR-335-5p in the 3′ UTR of RASA1 and associated luciferase assay. SW480 cells were co-transfected with either negative control (NC) or miR-335-5p mimics and luciferase reporter plasmids, including RASA1 3′ UTR WT or RASA1 3′ UTR mut. Data are presented as mean ± SD (n = 3). ∗p < 0.05. (D) Expression of RASA1 mRNA was determined by quantitative RT-PCR in SW480 and SW620 cells transfected with NC or miR-335-5p mimics. ∗∗p < 0.01. (E) Western blot analysis of RASA1 and Ras protein expression in SW480 and SW620 cells transfected with NC or miR-335-5p mimics. (F) Western blot analysis of E-cadherin and vimentin protein expression in SW480 and SW620 cells transfected with NC or miR-335-5p mimics. (G) Determination of the migration and invasion abilities of SW480 and SW620 cells transfected with NC or miR-335-5p mimics using a Transwell assay. Data are presented as mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01. Scale bars, 100 μm. (H) Determination of the migration and invasion abilities of SW480 and SW620 cells transfected with NC or miR-335-5p inhibitor using a Transwell assay. Data are presented as mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01. Scale bars, 100 μm.
To determine the effects of miR-335-5p on RASA1, miR-335-5p mimics were transiently transfected into SW480 and SW620 cells using Lipofectamine 3000. Evaluation with quantitative reverse-transcriptase PCR (qRT-PCR) and western blot showed that the mRNA and protein levels of RASA1 were significantly downregulated in the mimic-transfected cells (Figure 4D). In addition, miR-335-5p transfection increased expression of Ras protein (Figure 4E). Upregulation of miR-335-5p may be accompanied by EMT activation, as evidenced by the increased expression of vimentin and decreased expression of E-cadherin in SW480 and SW620 cells (Figure 4F). Moreover, we found that an increased level of miR-335-5p significantly promoted migration and invasion, whereas inhibition of miR-335-5p with a miR-335-5p inhibitor significantly suppressed migration and invasion of CRC cells (Figures 4G and 4H).
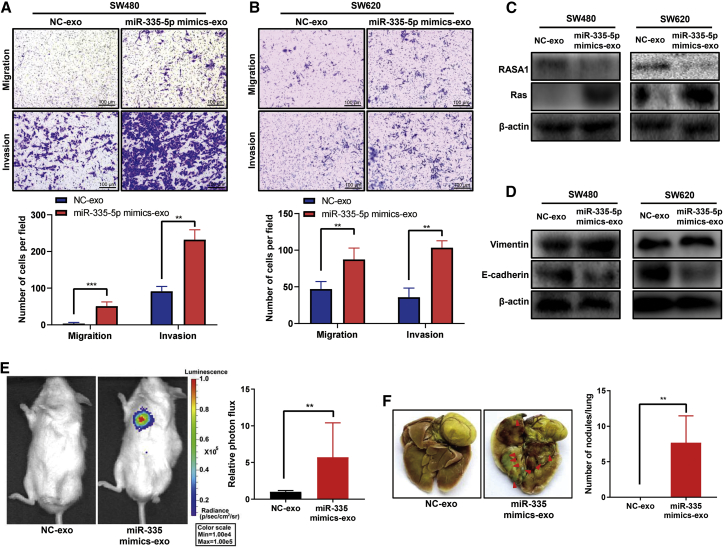
Exosome-transmitted miRNA-335-5p promotes CRC cell migration, invasion, EMT, and metastasis by targeting RASA1
Herein, we collected exosomes from the conditioned media of SW480 cells transfected with miR-335-5p mimic or a negative control (NC). The level of miR-335-5p in these exosomes was detected using qRT-PCR. The results showed miR-335-5p to be overexpressed in SW480-exo cells transfected with the mimic, indicating that miR-335-5p could be transferred from cells to their derived exosomes (Figure S5). We found that exosomes containing the mimic (miR-335-5p mimic-exo) significantly promoted migration and invasion of SW480 and SW620 cells (Figures 5A and 5B) and also decreased RASA1 protein expression and increased Ras protein expression (Figure 5C). Also, miR-335-5p mimic-exo suppressed EMT, as demonstrated by decreased expression of the epithelial marker E-cadherin and increased expression of the mesenchymal marker vimentin (Figure 5D). Finally, cells treated with miR-335-5p mimic-exo showed stronger promotion of metastasis in vivo (Figures 5E and 5F), with the number of macroscopic nodules on the lung surface dramatically increased. These results suggest that exosome-mediated shuttling of miR-335-5p may accelerate CRC cell migration and invasion in vitro and metastasis in vivo, which are accompanied by activation of Ras signaling and EMT.
Figure 5.
Exosome-transmitted miRNA-335-5p promotes CRC cell invasion, metastasis, and EMT, as well as decreases the level of RASA1
(A) Determination of the migration and invasion abilities of SW480 cells treated with NC-exo (SW480-exo transfected with NC miRNA) or miR-335-5p mimics-exo (SW480-exo transfected with miR-335-5p mimics) using a Transwell assay. Data are presented as mean ± SD (n = 3). ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 100 μm. (B) Determination of the migration and invasion abilities of SW620 cells treated with NC-exo or miR-335-5p mimics-exo using a Transwell assay. Data are presented as mean ± SD (n = 3). ∗∗p < 0.01. Scale bars, 100 μm. (C) Western blot analysis of RASA1 and Ras protein expression levels in SW480 and SW620 cells treated with NC-exo or miR-335-5p mimics-exo. (D) Western blot analysis of E-cadherin and vimentin protein expression levels in SW480 and SW620 cells treated with NC-exo or miR-335-5p mimics-exo. (E) Bioluminescence assay evaluating tumor metastases in mice. SW480 cells transduced with a lentiviral vector encoding the firefly luciferase gene (3 × 106 cells per mouse, n = 6) were injected into the tail vein of NOD/SCID mice, and the in vivo bioluminescent signal was quantified at the 9th week. Representative in vivo images of NC-exo or miR-335-5p mimics-exo treated mice are shown. Data are presented as mean ± SD (n = 6). ∗∗p < 0.01. (F) Representative photographs of macroscopic lung appearance. The number of metastatic tumor nodules for each group is shown on the right panel. Data are presented as mean ± SD (n = 6). ∗∗p < 0.01.
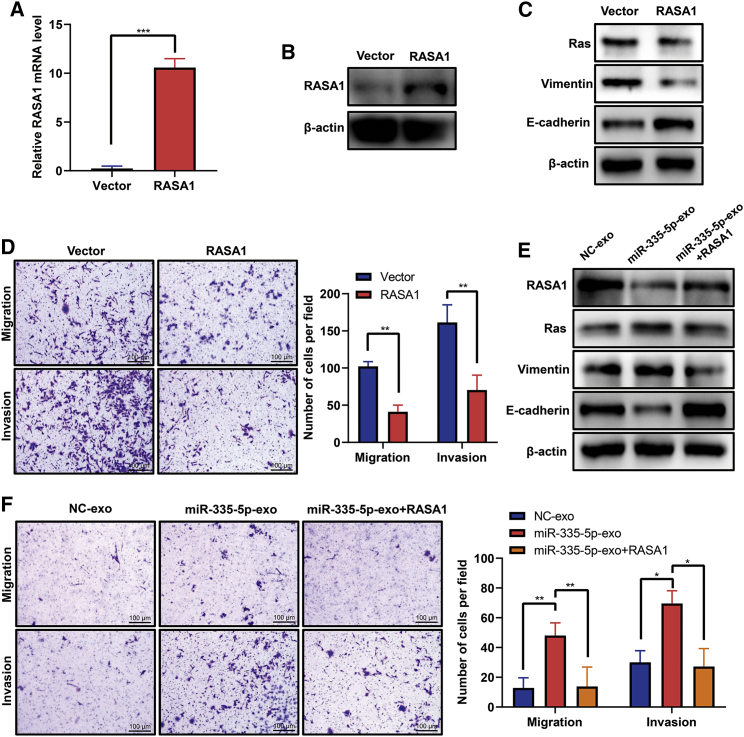
Upregulation of RASA1 eliminated the facilitation of exosomal miR-335-5p on migration and invasion of CRC cells
To further identify the potential function of RASA1 in CRC cells, we established a stable SW480 cell line overexpressing RASA1 using lentiviral vectors. As shown in Figures 6A and 6B, RASA1 mRNA and protein overexpression were, respectively, verified by real-time PCR and western blot in SW480 cells transfected with lentiviral vector expressing RASA1 (lenti-RASA1; SW480-RASA1). In cells overexpressing RASA1, protein levels of Ras and vimentin were significantly downregulated, whereas E-cadherin was upregulated (Figure 6C). Also, the migration and invasion abilities of SW480-RASA1 were significantly decreased compared with the SW480-vector control (Figure 6D).
Figure 6.
Ectopic expression of RASA1 inhibits CRC cell migration and invasion and eliminates the promotive effects of exosomal miR-335-5p on cell migration and invasion
(A) Quantitative RT-PCR showing an increased mRNA level of RASA1 in SW480 cells transduced with lentiviral vector expressing RASA1 (lenti-RASA1). Data are presented as mean ± SD (n = 3). ∗∗∗p < 0.001. (B) Western blot showing an increased protein level of RASA1 in SW480 cells transduced with lenti-RASA1. (C) Western blot showing decreased protein levels of Ras and vimentin and increased protein level of E-cadherin in SW480 cells transduced with lenti-RASA1. (D) Transwell migration and invasion assays of SW480 cells transduced with lenti-RASA1 or control vector. Representative images from triplicate experiments are shown. Data are presented as mean ± SD (n = 3). ∗p < 0.01. Scale bars, 100 μm. (E) Western blot analysis of RASA1, Ras, E-cadherin, and vimentin protein expression in SW480 cells treated with NC-exo, miR-335-5p mimics-exo, or miR-335-5p mimics-exo and lenti-RASA1. (F) Transwell migration and invasion assays of SW480 cells treated with NC-exo, miR-335-5p mimics-exo, or miR-335-5p mimics-exo and lenti-RASA1. Representative images from triplicate experiments are shown. Data are presented as mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01. Scale bars, 100 μm.
Next, we investigated whether RASA1 is involved in the induction of migration, invasion, and EMT by exosomal miR-335-5p in CRC cells. Western blot analysis revealed that miR-335-5p mimic-exo increased the protein levels of Ras and vimentin but decreased the protein levels of RASA1 and E-cadherin; with RASA1 overexpression, the inverse expression trends were observed for these proteins (Figure 6E). Moreover, RASA1 overexpression reversed miR-335-5p mimic-exo-induced migration and invasion of SW480 cells (Figure 6F). These results suggest that exosomal miR-335-5p might induce migration, invasion, and EMT by targeting RASA1.
Discussion
Metastasis and recurrence are the major causes of poor prognosis in patients suffering from CRC.25,26 Hence, a better understanding of the underlying mechanism of CRC metastasis and recurrence would provide a better strategy for suppressing CRC development, along with discovering novel regulators of these processes. The crosstalk between cancer cells is responsible for cancer cell metastasis, invasion, and other oncological behaviors.27,28 Dysregulation of miRNAs and exosomal shuttling of miRNAs may serve as critical factors in CRC progression and metastasis.29,30 Exosomes can effectively load and deliver miRNAs, causing some genes to be partially silenced in recipient cells, which in turn, affects metastasis-promoting tumor cell activities.23 In this study, we found that exosomal miR-335-5p derived from a metastatic CRC cell line (SW620) promotes CRC cell invasion and metastasis by activating EMT and RAS signaling via its target gene RASA1.
We extracted exosomes from the CRC cell lines SW480 (from primary lesions) and SW620 (from metastatic lesions) and evaluated their potential role in CRC. We showed that exosomes derived from the metastatic CRC cell line (SW620) promote CRC cell migration, invasion, and EMT. These findings suggest that the shuttling of exosomes and their cargo between CRC cells results in metastatic alterations, but the underlying mechanism of this process remained to be determined. Next, exosomal miRNAs from the two CRC cell lines were sequenced, and the potential miRNAs involved in metastasis in CRC were evaluated. Expression of miR-335-5p was found to be significantly increased in SW620-derived exosomes, which was in line with the qRT-PCR results. In addition, analysis of data downloaded from TCGA showed that high levels of miR-335-5p in CRC patients were associated with poor prognosis. More interestingly, GSEA analysis of mRNA profiles from CRC patients grouped according to miR-335-5p expression levels revealed that genes involved in the Ras protein signaling pathway and regulation of the Ras protein signaling pathway were significantly dysregulated. Notably, Ras signaling plays a crucial role in CRC progression.31
Previous cancer studies showed that miR-335-5p can be a promising invasive biomarker for cancer metastasis.32,33 For example, a high level of hsa-miR-335 was observed in patients with gastric cancer, who suffer from high-frequency recurrence and poor survival.32 Moreover, miR-335-5p was highly expressed in uterine sarcoma patients suffering from tumor metastasis and relapse, suggesting this miRNA can be a potential marker for prognosis and treatment response of uterine sarcomas.33 Our results revealed that upregulation of miR-335-5p positively promoted the invasive ability of CRC cells and its downregulation, the inverse. We additionally investigated the mechanism by which miR-335-5p mediates CRC cell metastasis. Luciferase reporter gene assays, qRT-PCR, and western blot analysis demonstrated that RASA1, a key tumor-suppressor gene, was the direct target of miR-335-5p. RASA1, which encodes p120 Ras GTPase activating protein (RasGAP), is known to be a negative regulator of the RAS/mitogen-activated protein kinase (MAPK) pathway; for example, downregulation of RASA1 in lung carcinoma cancer cells activated signaling downstream of RAS and promoted malignant transformation.34 As expected, RASA1 expression was significantly downregulated in CRC cells transfected with miR-335-5p mimics. Moreover, RASA1 downregulation in CRC cells promoted the expression of Ras and induced EMT. Furthermore, we also revealed that exosome-encapsulated miR-335-5p decreased the expression of RASA1, increased expression of Ras, and induced EMT and metastatic ability in recipient CRC cells. Meanwhile, overexpression of RASA1 in recipient CRC cells significantly reversed the effects of exosomal miR-335-5p. Hence, it is possible that exosomal miR-335-5p promotes CRC metastasis and activates EMT through directly targeting RASA1.
In our study, a total of 16 potential target genes of miR-335-5p were predicted by three bioinformatics algorithms: TargetScan, miRDB, and miRTarBase. Of the 16 potential target genes, in addition to RASA1, multiple target genes of miR-335-5p have been reported. For example, miR-335-5p has been reported to target Daam1 and ROCK1 to promote chondrogenesis of mesenchymal stem cells.35 miR-335 has also been reported to exert a metastatic inhibitory effect by targeting SP1,36 ROCK1,37 and ZEB2.38 However, our results showed that miR-335-5p promotes CRC metastasis and activates EMT through directly targeting RASA1. Shu et al.39 reported that miR-335 promoted growth and invasion by targeting DAAM1 in malignant astrocytoma cells. Some researchers have defined miR-335-5p as a tumor suppressor;36,38,40,41,47 however, others argued that miR-335-5p acted as an accelerator in CRC.42,43 Interestingly, Scarola et al.44 discovered that miR-335 controls cell proliferation and neoplastic transformation in a p53-dependent manner. Briefly, overexpression of miR-335 in tumor cells carrying the p53 WT allele decreased Rb1 protein and activated the p53 tumor-suppressor pathway to limit cell proliferation and neoplastic transformation but increased miR-335-induced hyperproliferation and neoplastic transformation in the absence of the p53 tumor-suppressor pathway. The two CRC cell lines (SW480 and SW620) used in this study have one thing in common: there is a G > A mutation in codon 273 of the p53 gene, resulting in an Arg > His substitution. We speculated that the functions of miR-335-5p in CRC cell lines are correlated with the mutations in the p53 gene. Nevertheless, further studies are needed to figure out the underlying mechanism and relationship of miR-335-5p with p53.
Evidence is accumulating that exosomal miRNAs facilitate invasive phenotypes in a number of cancers via various signaling pathways. Fang et al.45 revealed that miR-103, transferred by exosomes, can enhance vascular permeability and promotes metastasis through directly targeting junction proteins. Ultimately, the exosome-mediated transfer of miRNAs can alter target gene expression through post-transcriptional regulation and subsequently change the properties of recipient cells. However, there is no prior report that miRNAs specifically promote CRC cell metastasis through exosome-mediated cell-cell communication. Our results showed that miR-335-5p, delivered by exosomes, could promote CRC cell invasion, migration, and EMT transition. Moreover, exosomal miR-335-5p significantly increased the number of lung metastasis nodules in vivo. Based on these findings, we conclude that exosomal miR-335-5p could function as a regulatory factor to stimulate the metastatic potential of CRC cells in vitro and in vivo.
In summary, this study uncovered a novel function of exosomal miR-335-5p in CRC metastasis and expounds that exosome-encapsulated miR-335-5p promotes CRC cell invasion, metastasis, and EMT transition by directly targeting RASA1. These findings strongly suggest that exosomal miR-335-5p could be a prognostic biomarker and lead to valuable therapeutic strategies for addressing CRC metastasis.
Materials and methods
Cell culture
CRC cell lines SW480 and SW620 were purchased from the Chinese Academy of Sciences (Shanghai, China) and cultured in Leibovitz’s L-15 medium (Gibco, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (FBS) (Gibco). All cell lines were cultured at 37°C and 5% CO2 in a humidified incubator.
Exosome isolation and characterization
Exosomes were isolated from SW480 and SW620 cells. Briefly, cells were washed and incubated at 37°C and 5% CO2 for 48 h in medium with exosome-depleted FBS. FBS was depleted of exosomes prior to use by ultra-centrifugation at 110,000 × g overnight at 4°C. Cell culture supernatants were collected and centrifuged at 300 × g for 10 min to pellet the cells and then at 10,000 × g for 30 min at 4°C to further remove cells and debris. The supernatant was filtered through a 0.22-μm filter to remove micro-vesicles larger than 200 nm, and the filtered supernatant was ultra-centrifuged at 110,000 × g for 70 min at 4°C. To further eliminate contaminating protein, the raw exosome pellet was resuspended in PBS and another ultra-centrifugation performed. The exosome-enriched pellets were resuspended in a small volume of PBS or lysis buffer where indicated. The lysed sample was used for total protein measurement, and the intact exosomes in PBS were stored at −80°C for further experiments.
The morphology of SW480-exo and SW620-exo cells was imaged by a transmission electron microscope (FEI Tecnai Spirit Bio-TWIN). The size distribution of exosomes was determined using a Malvern Zetasizer Nano ZS90 (Malvern, UK), according to the manufacturer’s instructions. The exosome markers CD63 and TSG101 were detected by western blotting.
Western blot analysis
Total proteins of cells or exosomes were extracted using radio immunoprecipitation assay (RIPA) lysis buffer, supplemented with a protease inhibitor, and quantified by the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL, USA). Western blot analysis was performed as previously described.46 The following primary antibodies were used: anti-CD63 (cat. #ab217345; Abcam, Cambridge, UK), anti-TSG101 (cat. #ab125011; Abcam), anti-β-actin (cat. #ab179467; Abcam), anti-RASA1 (cat. #ab40677; Abcam), anti-Ras (cat. #ab52939; Abcam), anti-E-cadherin (cat. #610181; BD Biosciences), anti-vimentin (cat. #550513; BD Biosciences), and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; cat. #ma5-15738; Thermo Fisher Scientific, Waltham, MA, USA).
Internalization of exosomes
SW480-exo and SW620-exo cells were stained with PKH67 (Sigma), as previously described.8 The PKH67-labeled exosomes were incubated with SW480 cells in a six-well plate for 24 h. After incubation, cells were washed with PBS three times and then stained with 10 μM Dil at 37°C for 20 min. The cells were counterstained with 4′-6-diamidino-2-phenylindole (DAPI) and analyzed by fluorescent confocal microscopy (Leica Microsystems, Mannheim, Germany).
Small RNA sequencing
Small RNA sequencing was performed with SW480-exo and SW620-exo cell lines, according to standard procedures provided by Illumina, including the preparation of libraries and sequencing experiments. Small RNA sequencing libraries were first prepared with TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, CA, USA). The constructed libraries were sequenced using an Illumina HiSeq 2500 with a sequencing read length of 1 × 50 bp. Raw reads were processed with ACGT101-miR (LC Sciences, Houston, TX, USA) to remove adaptor dimers, junk, low-complexity sequences, common RNA families (rRNA, tRNA, small nuclear RNA [snRNA], and small nucleolar RNA [snoRNA]), and repeats. Student’s t test was used to evaluate the differential expression of miRNAs based on normalized deep-sequencing counts.
qRT-PCR analysis of miRNA expression
Total RNA of cells or exosomes was isolated using the RNeasy Mini Kit (QIAGEN; cat. #/ID: 74104), according to the manufacturer’s instructions. Total RNA (1 μg) was reverse transcribed into cDNA using the SuperScript First-Strand Synthesis Kit (Invitrogen, Thermo Fisher Scientific). Then, the cDNA was used for quantitative real-time PCR analysis performed on the QuantStudio 5 detection system (Applied Biosystems, Warrington, UK) using SYBR Premix EX Taq (Takara, Shiga, Japan) and miRNA-specific primers (Ribobio, Guangzhou, China). U6 was selected as the reference gene, and all reactions were performed in triplicate.
Colony-forming assay
Cells were dissociated and placed in a six-well plate at a density of 500 cells/well and then cultured in medium with 60 μg/mL SW480-exo or SW620-exo cells for 14 days (the medium was changed every 4 days). Clone-forming ability was assessed by staining with 0.1% crystal violet. All experiments were performed in triplicate.
Proliferation assay
Cells were dissociated and placed in a 96-well plate at 3 × 103 cells/well. SW480-exo and SW620-exo, quantified by total protein detection, were added to the culture medium at a final concentration of 60 μg/mL, and the plates were incubated at 37°C and 5% CO2 for the indicated times. Cell viability was assessed using the CCK-8 (Dojindo, Japan). All experiments were performed in triplicate.
Migration and invasion assays
Cell migration and invasion assays were performed using 24-well Transwell chamber inserts with polycarbonate membranes having a pore size of 8 μm (Corning Costar, Cambridge, MA, USA). The experiment was carried out according to the manufacturer’s instructions. Before the assays, cells were incubated with serum-free medium for 16 h at 37°C and 5% CO2. For the migration assay, 1 × 105 cells were resuspended in 200 μL serum-free medium and seeded in each insert and then incubated with 60 μg/mL SW480-exo or SW620-exo for 24 h. After incubation, cells were immobilized with paraformaldehyde for 20 min and stained with crystal violet for 25 min. The cells on the upper surface were wiped out with a cotton-tipped swab, and cells that migrated to the lower surface of the membrane were counted (five fields each chamber) using a microscope. For the invasion assay, the membranes were coated with Matrigel (BD Biosciences, the Netherlands) and dried at room temperature for 1 h before the assay. Subsequent experiments were similar to the migration assay.
Wound-healing assay
SW480 cells were seeded in a six-well plate and allowed to form monolayers overnight. A “scratch” was created using a 200-μL pipette tip, and the debris was removed. The cells were then treated with 60 μg/mL SW480-exo or SW620-exo for 48 h or with PBS as a control. Images were taken at 0, 24, and 48 h time points through a microscope (Leica, Germany) with the plate in the same position. All experiments were performed in triplicate.
Mining gene-expression data from TCGA
To identify the potential function of hsa-miR-335-5p in CRC, we downloaded miRNA and mRNA expression data for colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) from TCGA through the Genomic Data Commons (GDC) Data Transfer Tool. The miRNA expression data were obtained from 625 samples, consisting of 11 normal samples and 614 tumor samples. There were 608 matched pairs of miRNA and mRNA expression data, all of which were from tumor samples. The R package DESeq2 was used to quantify differential expression of miRNAs based on count data; the Kolmogorov-Smirnov1 test and Mann-Whitney U test were used to evaluate the degree of difference. K-means clustering was used to divide tumor samples into groups based on level of hsa-miR-335-5p expression—either high or low. GSEA was performed on the two groups.
The TargetScan algorithm (v7.0 context++) was used to predict miRNA binding sites. Only canonical 7−8 nt 3′ UTR sites were considered. Target sites with a context++ score ≤−0.4 were considered as high-confidence binding sites for analysis. Genes that were both targeted by hsa-miR-335-5p and had significant Spearman correlation coefficient (FDR < 0.05) were used in gene function enrichment analysis with the R package clusterProfiler.
Luciferase reporter gene assay
The WT or mut 3′ UTR of RASA1 was inserted into the psiCHECK-2 luciferase reporter vector (Promega, Madison, WI, USA) using specific primers (RASA1 3′ UTR-forward [F]-XhoI: 5′-CCGCTCGAGGATTCAGCATGTCCAACA-3′, RASA1 3′ UTR-reverse [R]-NotI: 5′-ATAAGAATGCGGCCGCTCTGCAATCCAGTTATACAC-3′). All constructed plasmids were verified by sequencing. SW480 cells were seeded in 24-well plates at a density of 5 × 104 and co-transfected with the psiCHECK-2 luciferase reporter vector containing RASA1 3′ UTR-WT or RASA1 3′ UTR-mut, along with a miRNA-335-5p mimic. Cell lysates were harvested 48 h post-transfection, after which, Dual-Luciferase Reporter Assay System (Promega) detection was carried out according to the instructions of the manufacturer. All reporter gene detections were repeated at least three times.
Lentiviral vector construction and transduction
A fragment of the RASA1 gene was amplified by PCR and inserted into a lentiviral vector as described previously.23 Lentiviral particles for transduction were produced by co-transfecting the transfer vector and three packaging vectors (pCMV-VSVG, pMDLg/pRRE, and pRSV-REV) into HEK293T cells for 48 h. Ultracentrifugation was subsequently performed to purify the harvested lentiviral particles. For transduction, 5 × 104 SW480 cells were seeded in a 24-well plate and transduced with lentivirus as previously described.23 The mRNA and protein levels of RASA1 were, respectively, detected by quantitative real-time PCR and western blotting.
Metastasis mouse model
A SW480 cell line expressing the luciferase gene was generated by transduction with a lentiviral vector (pLVX-GFP) encoding firefly luciferase. Transduction efficiency was checked with a fluorescence microscope at 72 h post-infection. To determine the role of exosomal miR-335-5p in the metastasis of CRC in vivo, luciferase-labeled SW480 cells (3 × 106) and 60 μg miR-335-5p mimic-exo or SW480-exo transfected with NC miRNA (NC-exo) were co-cultured for 24 h in vitro to achieve chemotactic progression to tumor metastatic cells. Then, these cells, with another 60 μg exosomes, were co-injected into mice via the tail vein. Briefly, 6- to 8-week-old male non-obese diabetic (NOD)-severe combined immunodeficiency (SCID) mice were purchased and randomly divided into two groups (six mice per group). One group was injected with luciferase-labeled SW480 cells, supplemented with NC-exo; the other was injected with luciferase-labeled SW480, supplemented with miR-335-5p mimic-exo. Noninvasive bioluminescence imaging was performed to detect the metastasis of CRC cells in vivo at 8 weeks post-injection. Luminescence images were recorded using an IVIS imaging machine, and the luminescence photon flux signal was calculated. All animal experiments were approved by the Animal Experimental Ethics Committee of Wenzhou Medical University (permit number: wydw2020-0824).
Statistical analysis
GraphPad Prism software (GraphPad Software, San Diego, CA, USA) was used for statistical analyses. Data are presented as mean ± SD. Student’s t test or Mann-Whitney test was used to calculate differences between groups. Survival rates were determined using the Kaplan-Meier method and the log-rank test. A p value of less than 0.05 was considered statistically significant.
Acknowledgments
This work was supported by the Natural Science Foundation of Zhejiang Province (grant number LY19H160024), National Natural Science Foundation of China (grant number 81672385), and Wenzhou Science and Technological Project (grant number Y20160159).
Author contributions
X.S., F.L., and L.J. conceived and designed the project and wrote the manuscript. X.S., F.L., W.S., W.Zhu., D.F., L.L., S.L., and W.Zhang. completed the experiments and analyzed the data. L.J. is responsible for research supervision and funding acquisition. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.02.022.
Supplemental information
References
- 1.Wolf A.M.D., Fontham E.T.H., Church T.R., Flowers C.R., Guerra C.E., LaMonte S.J., Etzioni R., McKenna M.T., Oeffinger K.C., Shih Y.T. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J. Clin. 2018;68:250–281. doi: 10.3322/caac.21457. [DOI] [PubMed] [Google Scholar]
- 2.Kopetz S., Chang G.J., Overman M.J., Eng C., Sargent D.J., Larson D.W., Grothey A., Vauthey J.N., Nagorney D.M., McWilliams R.R. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J. Clin. Oncol. 2009;27:3677–3683. doi: 10.1200/JCO.2008.20.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Christofori G. New signals from the invasive front. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A., Siegel R., Ward E., Murray T., Xu J., Smigal C., Thun M.J. Cancer statistics, 2006. CA Cancer J. Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 6.Juez I., Rubio C., Figueras J. Multidisciplinary approach of colorectal liver metastases. Clin. Transl. Oncol. 2011;13:721–727. doi: 10.1007/s12094-011-0722-x. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Gu Y., Cao X. The exosomes in tumor immunity. OncoImmunology. 2015;4:e1027472. doi: 10.1080/2162402X.2015.1027472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou W., Fong M.Y., Min Y., Somlo G., Liu L., Palomares M.R., Yu Y., Chow A., O’Connor S.T., Chin A.R. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melo S.A., Luecke L.B., Kahlert C., Fernandez A.F., Gammon S.T., Kaye J., LeBleu V.S., Mittendorf E.A., Weitz J., Rahbari N. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi M., Sawada K., Nakamura K., Yoshimura A., Miyamoto M., Shimizu A., Ishida K., Nakatsuka E., Kodama M., Hashimoto K. Exosomal miR-1290 is a potential biomarker of high-grade serous ovarian carcinoma and can discriminate patients from those with malignancies of other histological types. J. Ovarian Res. 2018;11:81. doi: 10.1186/s13048-018-0458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B., Pan X., Cobb G.P., Anderson T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H., Li Y., Lai M. The microRNA network and tumor metastasis. Oncogene. 2010;29:937–948. doi: 10.1038/onc.2009.406. [DOI] [PubMed] [Google Scholar]
- 17.Valastyan S., Reinhardt F., Benaich N., Calogrias D., Szász A.M., Wang Z.C., Brock J.E., Richardson A.L., Weinberg R.A. Retraction notice: A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2015;161:417. doi: 10.1016/j.cell.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory P.A., Bracken C.P., Bert A.G., Goodall G.J. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z., Li Y., Ahmad A., Azmi A.S., Kong D., Banerjee S., Sarkar F.H. Targeting miRNAs involved in cancer stem cell and EMT regulation: An emerging concept in overcoming drug resistance. Drug Resist. Updat. 2010;13:109–118. doi: 10.1016/j.drup.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simons M., Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Redis R.S., Calin S., Yang Y., You M.J., Calin G.A. Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol. Ther. 2012;136:169–174. doi: 10.1016/j.pharmthera.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao L.Q., Yang X.W., Chen Y.B., Zhang D.W., Jiang X.F., Xue P. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol. Cancer. 2019;18:148. doi: 10.1186/s12943-019-1075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyuno D., Zhao K., Bauer N., Ryschich E., Zöller M. Therapeutic Targeting Cancer-Initiating Cell Markers by Exosome miRNA: Efficacy and Functional Consequences Exemplified for claudin7 and EpCAM. Transl. Oncol. 2019;12:191–199. doi: 10.1016/j.tranon.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kogure A., Kosaka N., Ochiya T. Cross-talk between cancer cells and their neighbors via miRNA in extracellular vesicles: an emerging player in cancer metastasis. J. Biomed. Sci. 2019;26:7. doi: 10.1186/s12929-019-0500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H., Wang C., Song H., Xu Y., Ji G. RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol. Cancer. 2019;18:8. doi: 10.1186/s12943-018-0932-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardhan K., Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers (Basel) 2013;5:676–713. doi: 10.3390/cancers5020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erdogan B., Webb D.J. Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochem. Soc. Trans. 2017;45:229–236. doi: 10.1042/BST20160387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neri S., Ishii G., Hashimoto H., Kuwata T., Nagai K., Date H., Ochiai A. Podoplanin-expressing cancer-associated fibroblasts lead and enhance the local invasion of cancer cells in lung adenocarcinoma. Int. J. Cancer. 2015;137:784–796. doi: 10.1002/ijc.29464. [DOI] [PubMed] [Google Scholar]
- 29.Qu A., Yang Y., Zhang X., Wang W., Liu Y., Zheng G., Du L., Wang C. Development of a preoperative prediction nomogram for lymph node metastasis in colorectal cancer based on a novel serum miRNA signature and CT scans. EBioMedicine. 2018;37:125–133. doi: 10.1016/j.ebiom.2018.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L., Zhang S., Yao J., Lowery F.J., Zhang Q., Huang W.C., Li P., Li M., Wang X., Zhang C. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100–104. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koveitypour Z., Panahi F., Vakilian M., Peymani M., Seyed Forootan F., Nasr Esfahani M.H., Ghaedi K. Signaling pathways involved in colorectal cancer progression. Cell Biosci. 2019;9:97. doi: 10.1186/s13578-019-0361-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan Z., Xiong Y., Xu W., Gao J., Cheng Y., Wang Z., Chen F., Zheng G. Identification of hsa-miR-335 as a prognostic signature in gastric cancer. PLoS ONE. 2012;7:e40037. doi: 10.1371/journal.pone.0040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez Dos Anjos L., de Almeida B.C., Gomes de Almeida T., Mourão Lavorato Rocha A., De Nardo Maffazioli G., Soares F.A., Werneck da Cunha I., Baracat E.C., Carvalho K.C. Could miRNA Signatures be Useful for Predicting Uterine Sarcoma and Carcinosarcoma Prognosis and Treatment? Cancers (Basel) 2018;10:315. doi: 10.3390/cancers10090315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi T., Desmeules P., Smith R.S., Drilon A., Somwar R., Ladanyi M. RASA1 and NF1 are Preferentially Co-Mutated and Define A Distinct Genetic Subset of Smoking-Associated Non-Small Cell Lung Carcinomas Sensitive to MEK Inhibition. Clin. Cancer Res. 2018;24:1436–1447. doi: 10.1158/1078-0432.CCR-17-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin X., Wu L., Zhang Z., Yang R., Guan Q., Hou X., Wu Q. MiR-335-5p promotes chondrogenesis in mouse mesenchymal stem cells and is regulated through two positive feedback loops. J. Bone Miner. Res. 2014;29:1575–1585. doi: 10.1002/jbmr.2163. [DOI] [PubMed] [Google Scholar]
- 36.Xu Y., Zhao F., Wang Z., Song Y., Luo Y., Zhang X., Jiang L., Sun Z., Miao Z., Xu H. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2012;31:1398–1407. doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Zhao W., Fu Q. miR-335 suppresses migration and invasion by targeting ROCK1 in osteosarcoma cells. Mol. Cell. Biochem. 2013;384:105–111. doi: 10.1007/s11010-013-1786-4. [DOI] [PubMed] [Google Scholar]
- 38.Sun Z., Zhang Z., Liu Z., Qiu B., Liu K., Dong G. MicroRNA-335 inhibits invasion and metastasis of colorectal cancer by targeting ZEB2. Med. Oncol. 2014;31:982. doi: 10.1007/s12032-014-0982-8. [DOI] [PubMed] [Google Scholar]
- 39.Shu M., Zheng X., Wu S., Lu H., Leng T., Zhu W., Zhou Y., Ou Y., Lin X., Lin Y. Targeting oncogenic miR-335 inhibits growth and invasion of malignant astrocytoma cells. Mol. Cancer. 2011;10:59. doi: 10.1186/1476-4598-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo L., Xia L., Zha B., Zuo C., Deng D., Chen M., Hu L., He Y., Dai F., Wu J. miR-335-5p targeting ICAM-1 inhibits invasion and metastasis of thyroid cancer cells. Biomed. Pharmacother. 2018;106:983–990. doi: 10.1016/j.biopha.2018.07.046. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y., Wang F., Huang H., Zhang Y., Xie H., Men T. lncRNA SLCO4A1-AS1 promotes growth and invasion of bladder cancer through sponging miR-335-5p to upregulate OCT4. OncoTargets Ther. 2019;12:1351–1358. doi: 10.2147/OTT.S191740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Y., Yang H., Yuan L., Liu G., Zhang C., Hong M., Liu Y., Zhou M., Chen F., Li X. Overexpression of miR-335 confers cell proliferation and tumour growth to colorectal carcinoma cells. Mol. Cell. Biochem. 2016;412:235–245. doi: 10.1007/s11010-015-2630-9. [DOI] [PubMed] [Google Scholar]
- 43.Vickers M.M., Bar J., Gorn-Hondermann I., Yarom N., Daneshmand M., Hanson J.E., Addison C.L., Asmis T.R., Jonker D.J., Maroun J. Stage-dependent differential expression of microRNAs in colorectal cancer: potential role as markers of metastatic disease. Clin. Exp. Metastasis. 2012;29:123–132. doi: 10.1007/s10585-011-9435-3. [DOI] [PubMed] [Google Scholar]
- 44.Scarola M., Schoeftner S., Schneider C., Benetti R. miR-335 directly targets Rb1 (pRb/p105) in a proximal connection to p53-dependent stress response. Cancer Res. 2010;70:6925–6933. doi: 10.1158/0008-5472.CAN-10-0141. [DOI] [PubMed] [Google Scholar]
- 45.Fang J.H., Zhang Z.J., Shang L.R., Luo Y.W., Lin Y.F., Yuan Y., Zhuang S.M. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. 2018;68:1459–1475. doi: 10.1002/hep.29920. [DOI] [PubMed] [Google Scholar]
- 46.Jiang L., Chen Y., Chan C.Y., Wang X., Lin L., He M.L., Lin M.C., Yew D.T., Sung J.J., Li J.C., Kung H.F. Down-regulation of stathmin is required for TGF-beta inducible early gene 1 induced growth inhibition of pancreatic cancer cells. Cancer Lett. 2009;274:101–108. doi: 10.1016/j.canlet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 47.An Y., Cai H., Zhang Y., Liu S., Duan Y., Sun D., Chen X., He X. circZMYM2 Competed Endogenously with miR-335-5p to Regulate JMJD2C in Pancreatic Cancer. Cell. Physiol. Biochem. 2018;51:2224–2236. doi: 10.1159/000495868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.