Figure 1.
Establishing specificity groups with CDR3β sequences from lung cancer patients
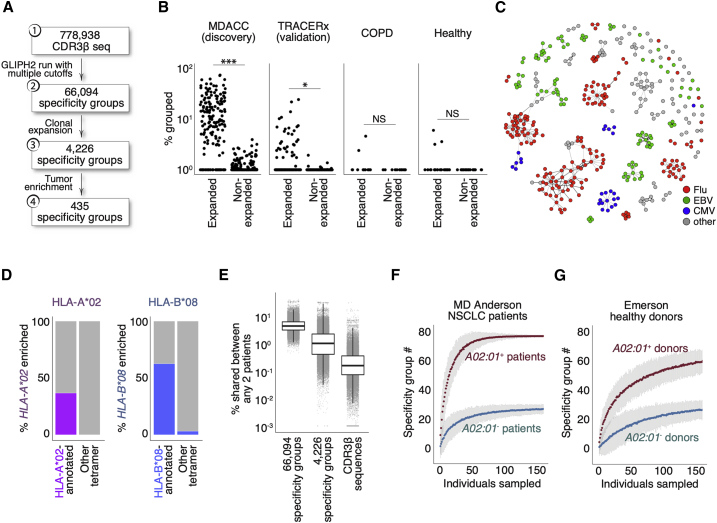
(A) Analysis of shared T cell specificities with the GLIPH2 algorithm. Step 1: 778,938 CDR3β sequences from the MDACC cohort as input for GLIPH2 analysis. Step 2: establish 66,094 specificity groups with multiple criteria (Figure S1A). Step 3: establish 4,226 clonally expanded specificity groups. Step 4: establish 435 clonally expanded, tumor-enriched specificity groups.
(B) Clinical relevance of tumor-enriched specificity groups in lung cancer. The most clonally expanded CDR3β sequences from tumors belonged to the 435 tumor-enriched specificity groups, whereas those from lung tissues of healthy donors and COPD patients did not. The trend was validated with tumors from a second NSCLC cohort (the TRACERx consortium, n = 202, validation). ∗∗∗p < 0.001; ∗p < 0.05 by paired t test. NS, not significantly different.
(C) Network analysis of 396 specificity groups annotated with CDR3β sequences from HLA tetramers with flu (red), EBV (green), and CMV (blue) antigens. Each dot is a specificity group, edges indicate the presence of identical CDR3β sequence(s) shared across two specificity groups.
(D) Percentage (%) of HLA-A∗02 or HLA-B∗08 tetramer-annotated specificity groups with significantly enriched the A∗02 (purple, left plot) or B∗08 (blue, right plot) supertype alleles, respectively. Specificity groups annotated with tetramers of other HLA alleles (other tetramer) were included for comparisons.
(E) Percentage of shared specificity between any two given MDACC NSCLC patients (% shared between any 2 patients, total n = 178) based on CDR3β membership in total specificity groups regardless of clonal expansion (n = 66,094), membership in clonally expanded specificity groups (n = 4,226), or comparison of identical CDR3β sequences. Boxes represent medians with the first (25th) and third (75th) quartiles.
(F and G) Bootstrapping of specificity group numbers (y axis, specificity group #) with varying sampling sizes (individuals sampled) for either HLA-A∗02+ or HLA-A∗02− NSCLC patients (F) or healthy donors (G, Emerson study). Data represent means with 3× standard errors from repeated sampling.