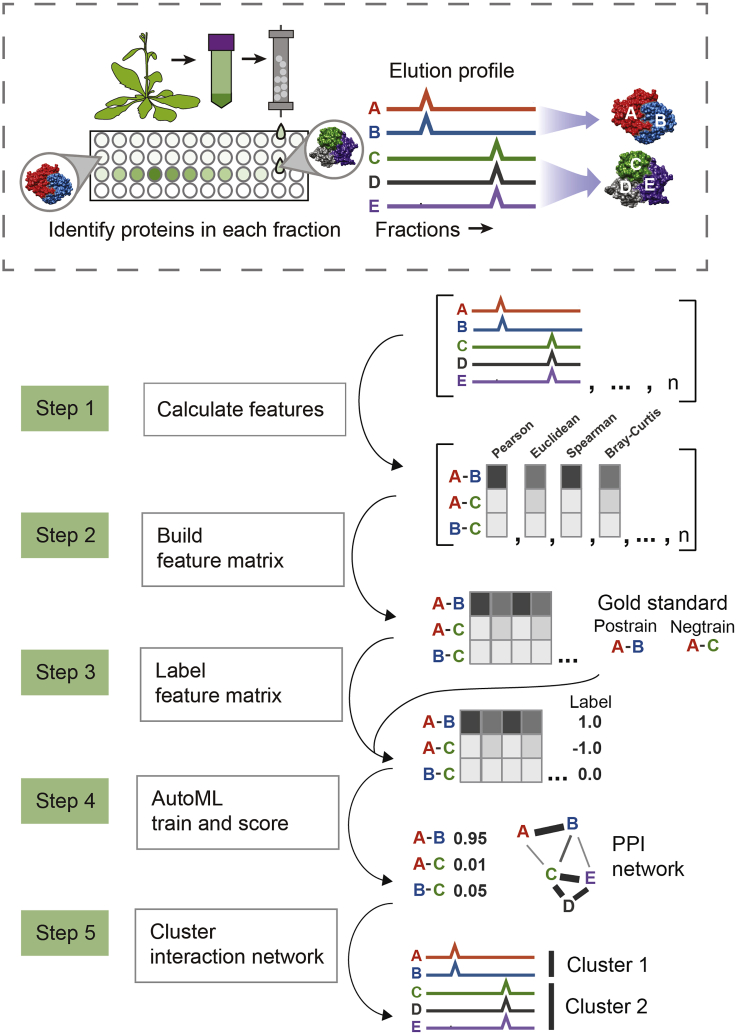
Figure 3.
Overview of computational pipeline to detect protein-protein interactions and protein complexes
In step 1, a set of similarity scores between all proteins are calculated for each fractionation experiment. In step 2, these similarity scores are combined into one large table. In step 3, pairs of proteins that are known from prior literature to interact are labeled with a 1 (positive training label), and a set of random pairs of proteins are labeled with a −1 (negative training label). In step 4, a model is trained to distinguish these positive and negatively labeled pairs of proteins, giving a score to each pair, where a higher score indicates higher probability of interaction. In step 5, this interaction network is clustered to protein complexes.