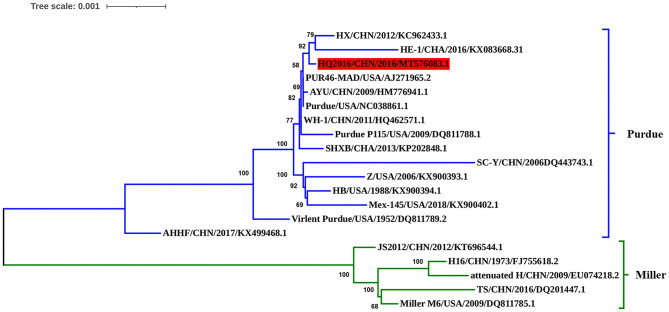
Figure 4.
Phylogenetic analysis of the complete genome sequences of the strain HQ2016, other TGEV reference strains. TGEV HQ2016 belongs to the Purdue cluster of TGEV, not the Miller cluster. Complete genome were aligned used Clustal W program which have trimed both 3′ and 5′ ends gaps between TGEV genomes Phylogenetic tree was constructed using the neighbor-joining method with the MEGA 6.0 program. The optimal tree with the sum of branch length = 0.02540989 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Tajima-Nei method.