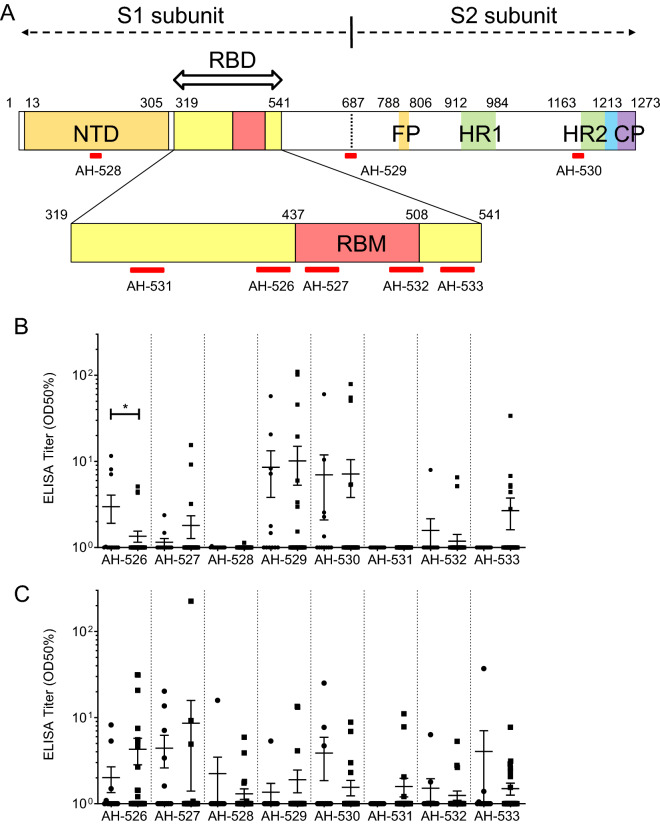
Figure 3.
Linear B cell epitopes on Spike protein of SARS-CoV-2 determined by ELISA. (A) Scheme of predicted linear B cell epitopes in the RBD or other regions of Spike protein in SARS-CoV-2. As shown in red bars, five regions (AH-531, 346–365 aa.; AH-526, 413–432 aa.; AH-527, 442–460 aa.; AH-532, 491–509 aa.; and AH-533, 518–537 aa.) were selected from the RBD, and three regions (AH-528, 146–164 aa.; AH-529, 671–690 aa.; and AH-530, 1146–1164 aa.) were selected from the NTD, S1 and S2 subunits. NTD, N-terminal domain; RBD, receptor-binding domain; RBM, receptor-binding motif; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; CP, cytoplasm domain. (B,C) The serum titer against synthetic SARS-CoV-2 peptides (AH-526 to AH-533) is expressed as the half-maximal binding (OD 50%). (B) Total IgG and (C) IgM. closed circle, serum samples collected from patients in the ICU of Osaka University Hospital (n = 12); closed square, serum samples collected from patients in Osaka City Juso Hospital (n = 31). All the data are expressed as the mean ± SEM. Statistical evaluation was performed by Mann–Whitney U test; *p < 0.05. Graphs made in GraphPad Prism version 8.4.3. https://www.graphpad.com/scientific-software/prism/.