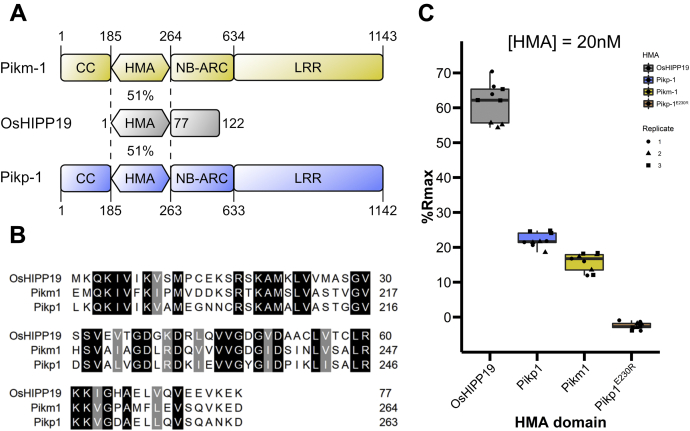
Figure 3.
AVR-PikD interacts with the HMA domain of OsHIPP19 with higher affinity than with the integrated HMA domains of Pikp-1 or Pikm-1.A, schematic representation of Pikm-1, Pikp-1 and OsHIPP19. OsHIPP19-HMA shares 51% sequence identity with both Pikp-HMA and Pikm-HMA. B, amino acid sequence alignment of the HMA domains of OsHIPP19, Pikp-1, and Pikm-1. The alignment was produced with Clustal Omega and colored using BoxShade. C, boxplots showing the %Rmax observed for the interactions between AVR-PikD and each of the HMA domains. %Rmax is the percentage of the theoretical maximum response, assuming a 1:1 HMA:effector binding model for OsHIPP19-HMA and Pikm-HMA, and a 2:1 binding model for Pikp-HMA and PikpE230R-HMA. The center line of the box represents the median and the box limits are the upper and lower quartiles. The whiskers extend to the smallest value within Q1 − 1.5× the interquartile range (IQR) and the largest value within Q3 + 1.5× IQR. Individual data points are represented as black shapes. The experiment was repeated three times, with each experiment consisting of three technical replicates. Plots were produced using the ggplot2 package (71) in R (72).