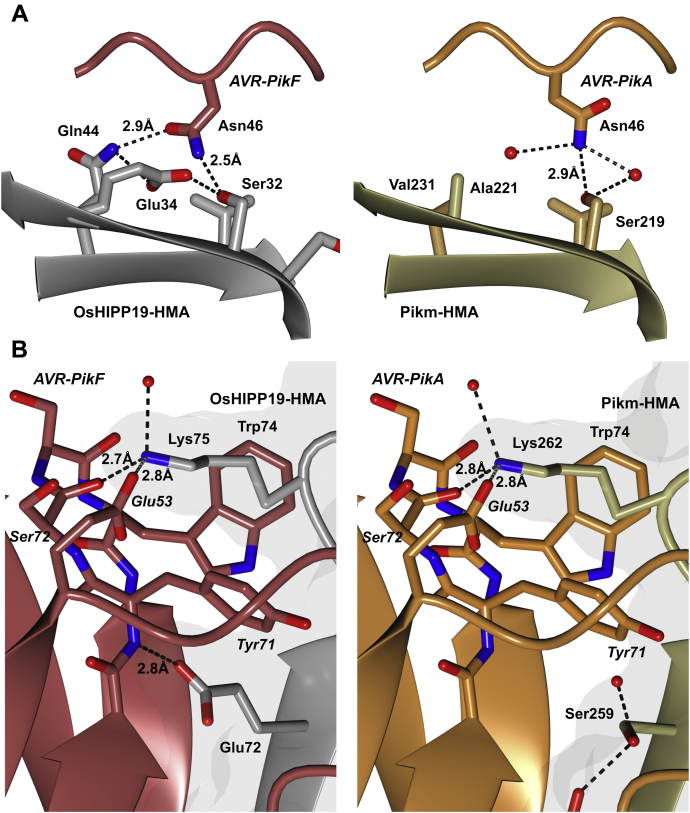
Figure 7.
Comparison of the binding interfaces in the crystal structure of AVR-PikF in complex with the HMA domain of OsHIPP19 (7B1I) with the previously published crystal structure of AVR-PikA in complex with the HMA domain of Pikm-1 (6FUD, (42)). The structures of AVR-PikF and OsHIPP19-HMA are represented as red and gray ribbons, respectively, with the molecular surface of OsHIPP19-HMA also shown. The structures of AVR-PikA and Pikm-HMA are represented as orange and gold ribbons, respectively, with the molecular surface of Pikm-HMA also shown. The side chains of amino acids of interest are displayed as cylinders. Hydrogen bonds are represented by dashed lines. A, AVR-PikFAsn46 is rotated and forms an additional hydrogen bond with OsHIPP19-HMA compared with AVR-PikAAsn46 bound to Pikm-HMA. The side chain of OsHIPP19Glu34 exists in two alternate conformations, both supported by the electron density. For clarity, only the relevant conformation is shown here. B, OsHIPP19Glu72 forms an additional hydrogen bond with the main chain of AVR-PikF.