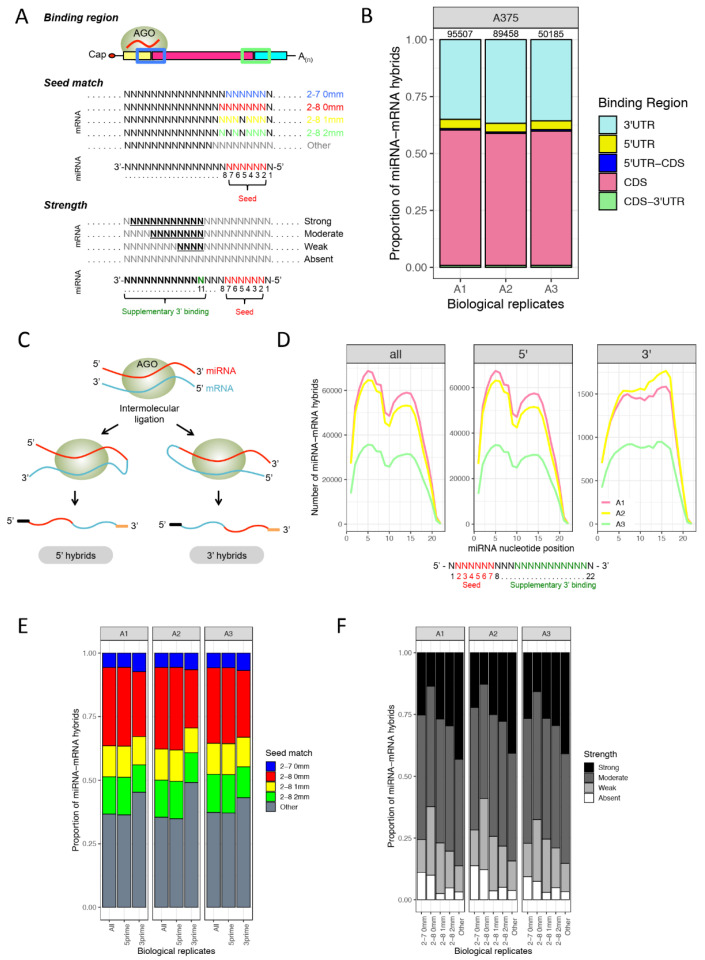
Figure 2.
Characteristics of miRNA-mRNA hybrids discovered in melanoma cells. (A) Binding characteristics of miRNA–mRNA interactions. Binding region: mRNA part of the hybrids mapped to the ENSEMBL transcript database with regions corresponding to the 5′UTR and 3′UTR, CDS, 5′-UTR-CDS, and CDS-3′UTR. Seed match: nucleotides (nt) 2–7 at 5′ end of miRNA with zero (2–7, 0 mm), 2–8 with zero (2–8, 0 mm), one (2–8, 1 mm), or two mismatches (2–8, 2 mm), and the remaining binding modes were classified as “other”. Seed pairing at nucleotides 2–7 (blue) and 2–8 (red) without mismatches was considered as canonical seed pairing, whereas any additional type of pairing was referred to as non-canonical seed pairing. Strength: “strength” of binding outside the seed region at the 3′ end of the miRNA based on number of bound nt: >8 nt (strong), 5–8 nt (moderate), 1–4 nt (weak), and 0 nt (absent). (B) Proportion of miRNA-mRNA hybrids across three biological replicates of the BRAF-mutant melanoma cell line A375 (A1, A2, and A3) with miRNA binding sites mapped to the mRNA sequence, and the number of identified hybrid sequences in each replicate. Hybrid mRNAs for which the transcript was not annotated in the corresponding database were excluded. The number above the bars displays the number of hybrids identified in each biological replicate. (C) The two types of miRNA-mRNA hybrid molecules that were generated during the intermolecular ligation: 5′ and 3′ miRNA hybrids, in which the miRNA is located at the 5′ end or the 3′ end of the hybrids, respectively. (D) M-plots showing the number of miRNA-mRNA hybrids bound at each miRNA nt position (x-axis, nt 1–22), and showing that most genes are targeted via the seed sequence but also with supplementary sites in the 3′ region of the miRNA. (E) The type of seed pairing (5′ miRNA sequence) was divided into classes as shown in (A). The proportion of miRNA-mRNA hybrids across the three biological replicates was plotted for the different seed categories for all hybrids as well as for 5′ and 3′ hybrids separately. (F) miRNA base-pairing via the 3′ portion of the miRNA (supplementary 3′ or non-seed pairing). The strength of 3′ sequence binding was classified as in (A) and was plotted for each seed sequence type across the biological replicates.