Figure 2: Effect of false read binning.
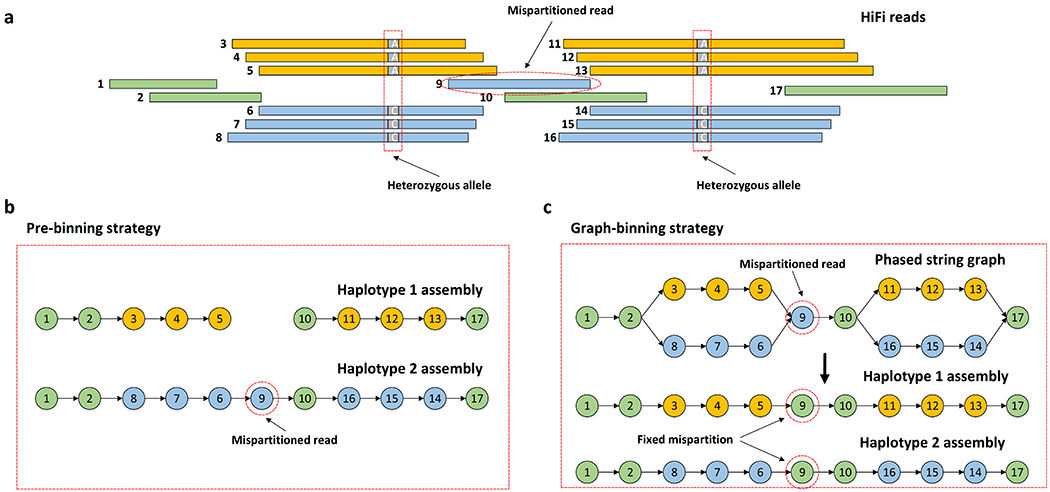
(a) A set of reads with global phasing information provided by the complementary data. Reads in orange and reads in blue are specifically partitioned into haplotype 1 and haplotype 2, respectively. The remaining reads in green are partitioned into both haplotypes. Read 9 without heterozygous alleles is mispartitioned into haplotype 2, instead of to both haplotypes. (b) Pre-binning assemblies produced by current methods which independently assemble two haplotypes. Haplotype 1 is broken into two contigs due to the mispartition of read 9. (c) Hifiasm fixes the mispartition by the local phasing information in the phased assembly graph. It is able to identify that read 9 does not have heterozygous alleles, so that read 9 should be partitioned into both haplotypes.
