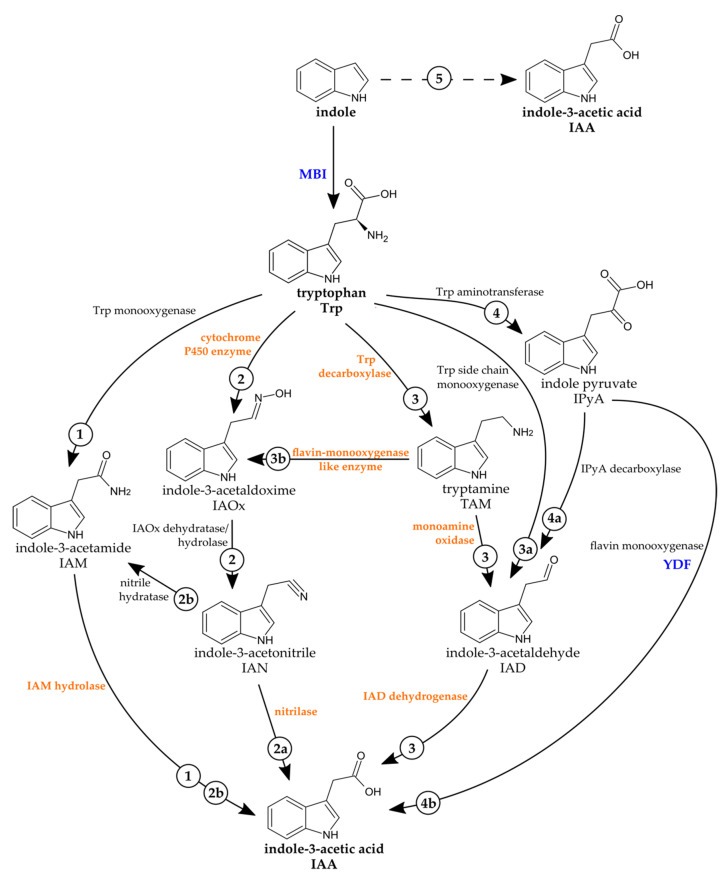
Figure 1.
Indole-3-acetic acid (IAA) biosynthesis pathways in bacteria, fungi, and plants with the known enzymes (modified after KEGG [36,37,38]). IAA can be synthesized from tryptophan (Trp-dependent pathways, 1–4) and directly from indole (Trp-independent pathways, 5). Plants synthesize IAA over the indole-3-acetamide (IAM) (1), the indole-3-acetaldoxime/indole-3-acetonitrile (IAOx–IAN) (2-2a), the tryptamine (TAM) (3-3b-2-2a), and the indole-3-pyruvic acid (IPyA) pathway (4-4a-3 or 4-4b). Bacteria and fungi prefer the IAM (1), the IAOx–IAN (2-2a or 2-2b), the TAM (3), or the IPyA (4-4a-3) pathway. The IAA pathway with the direct conversion of Trp to indole-3-acetaldehyde (IAD, without IPyA as intermediate, 3a-3) is only known in Pseudomonas species [39,40,41,42]. The reaction site of the IAA biosynthesis inhibitors 2-mercaptobenzimidazole (MBI) (Trp synthase) and yucasin DF (YDF) (YUC enzymes) is shown in blue. Enzymes for which candidate genes were found in C. asteris are highlighted in orange (see also Table 1).