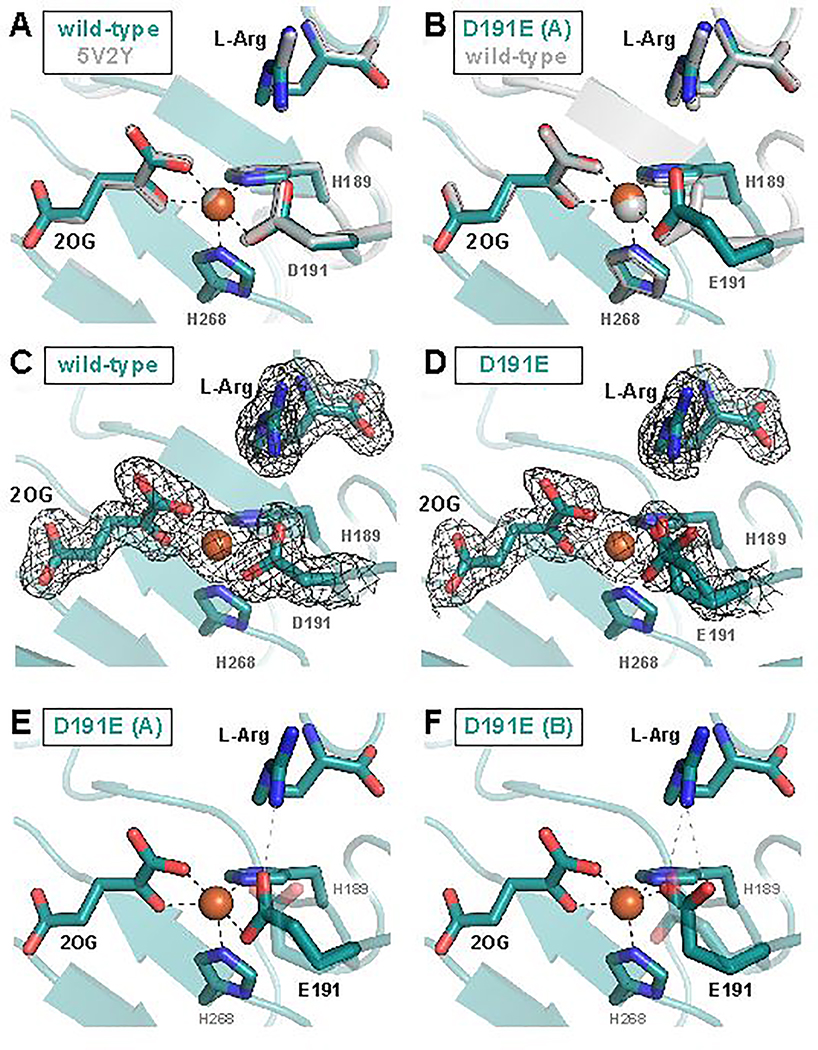
Figure 4.
X-ray crystallographic models of the EFE active site with 2OG and L-Arg bound. (A) Comparison of the 1.83-Å-resolution wild-type EFE•Fe•2OG•L-Arg structure (PDB accession code 6VP4) to the published 1.43-Å-resolution EFE•Mn•2OG•L-Arg structure (PDB accession code 5V2Y) reveals no significant differences. (B) One of two conformations of the carboxylate side chain observed in the D191E EFE•Fe•2OG•L-Arg structure (conformation A) closely resembles that observed in the wild-type enzyme. In (C) and (D), 2Fo-Fc maps contoured to 1.0σ are shown in black for wild-type and D191E EFE, respectively. Conformation A of the E191 side chain forms a single hydrogen bond with the L-Arg substrate (E), whereas Conformation B forms two hydrogen bonds (F).