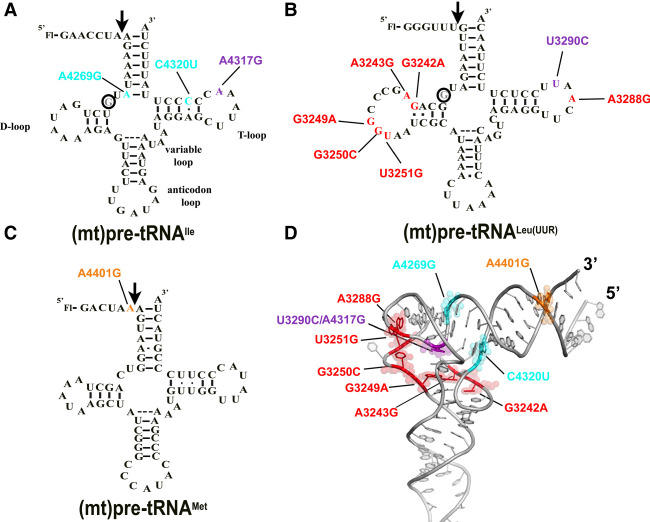
FIGURE 1.
Locations of the investigated mitochondrial tRNA mutations in the predicted secondary structure of (A) (mt)pre-tRNAIle, (B) (mt)pre-tRNALeu(UUR), and (C) (mt)pre-tRNAMet. Circles indicate methylation sites for the MRPP1/2 subcomplex. (D) Location of the mutations in the L-shaped tertiary structure of tRNAs. High-resolution structure of the investigated tRNAs are not available, therefore we used a conserved eukaryotic tRNAPhe with known structure (PDB ID: 1ehz) for generating the figure. Mutations are color coded according to the colors shown in A, B, and C. Purple represents mutation at the same position in (mt)pre-tRNALeu(UUR) (U3290C) and (mt)pre-tRNAIle (A4317G). We also note that the D-loop for (mt)pre-tRNALeu(UUR) and (mt)pre-tRNAMet is different in length from most canonical tRNAs and therefore the elbow region may fold differently than modeled here.