FIGURE 2.
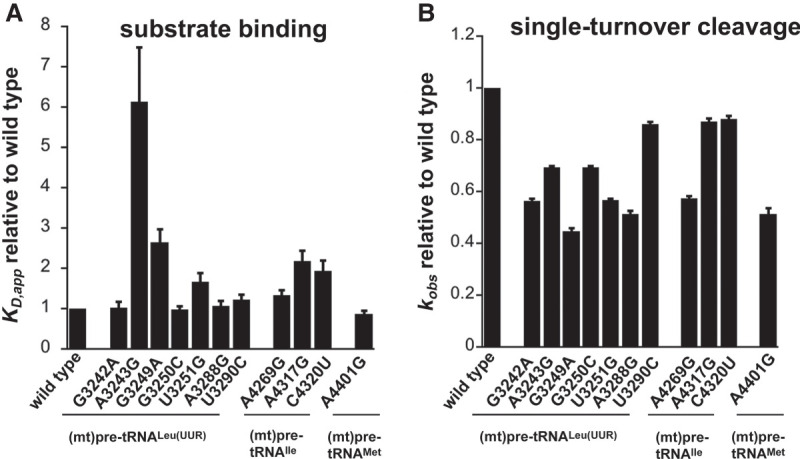
Disease-related tRNA mutations affect binding affinity and 5′ end pre-tRNA processing activity by the mtRNase P. (A) Binding affinities of mutant mitochondrial pre-tRNAs for mtRNase P using standard binding conditions. 20 nM fluorescently labeled substrates were preincubated with 150 nM MRPP1/2 for 5 min and then titrated with 0–5 µM MRPP3 as described in Karasik et al. (2019). Data were collected when binding reached equilibria from changes in fluorescence anisotropy. The KD values are calculated from the fit of the binding isotherm to the data and reported values are divided by the wild-type KD values. (B) Single-turnover cleavage rates of mitochondrial pre-tRNAs containing mutations catalyzed by mtRNase P using standard cleavage conditions. Reactions contained 20 nM fluorescently labeled substrate, 1 µM MRPP3, and 0.4 µM MRPP1/2. The kobs,cleavage values are calculated from the fit of the single exponential to the data and reported values are divided by the wild-type kobs,cleavage values.
