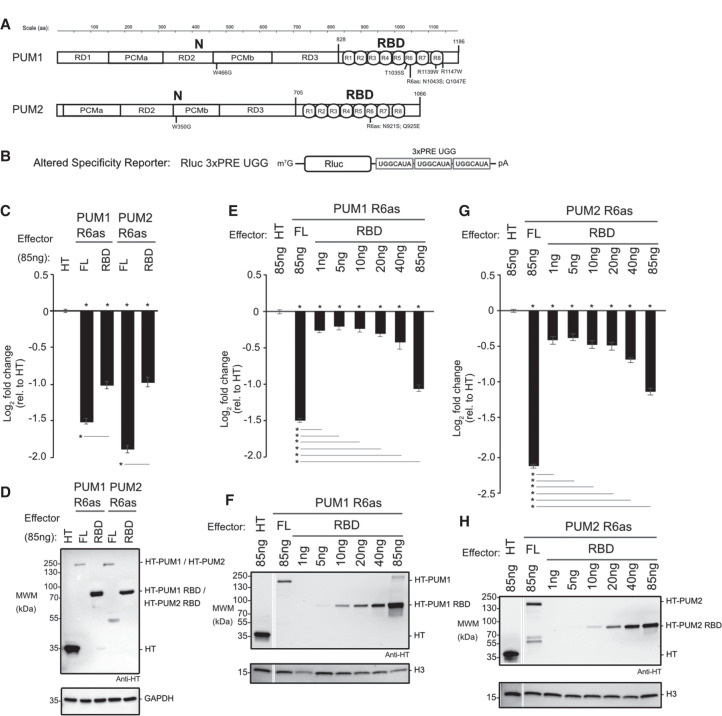
FIGURE 3.
The RNA-binding domains of PUM1&2 are necessary but not sufficient for repression. Mean log2 fold change values ±SEM are plotted and listed in Supplemental Table S1. (*) P-value <0.05 relative to the negative control Halotag (HT) (above x-axis) or between the indicated proteins (below the x-axis). (A) Domain organization of PUM1 and PUM2 indicating the amino-terminal region (N) with putative repression domains (RD), Pumilio conserved motifs (PCMa and PCMb), and RNA-binding domain (RBD) with eight PUF repeats (R1-8), as previously designated (Weidmann and Goldstrohm 2012; Goldstrohm et al. 2018). Residue numbering on top. Putative cap binding residues, W466 and W350, are indicated below (Cao et al. 2010). The residues of the sixth PUF repeat that were changed in the altered specificity constructs, PUM1 R6as and PUM2 R6as, are shown below. The PUM1 residues linked to the diseases PADDAS, R1139W and R1147W, and PRCA, T1035S, are indicated below. (B) Altered specificity assay reporter gene, Nluc 3xPRE UGG, that specifically measures the activity of exogenous PUM1&2 (R6as) programmed to bind to the UGG motifs, relative to negative control Halotag (HT). n = 9. (C) Altered specificity reporter assay comparing the repressive activity of full length (FL) or RBD PUM1 or PUM2 R6as effectors, relative to negative control HT. n = 9. (D) Western blot of HT-tagged proteins from a representative experimental replicate from samples from C. GAPDH served as loading control. (E) Comparison of RBD to full length (FL) PUM1 (R6as) using the altered specificity reporter assay. Total mass of transfected DNA was balanced across transfections with an empty vector. n = 9. (F) Western blot of HT-tagged PUM1 proteins in E from a representative experimental replicate. Histone H3 served as loading control. (G) Same as E, except using PUM2 FL and RBD (R6as) proteins. n = 9. (H) Western blot of HT-tagged PUM2 proteins used in G from a representative experimental replicate.