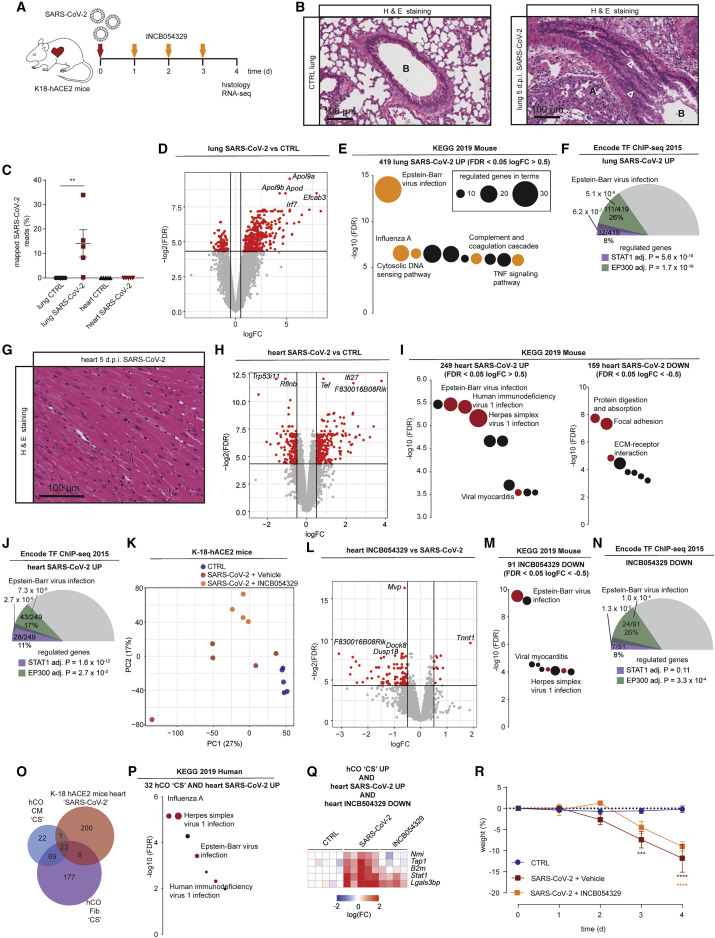
Figure 5.
SARS-CoV-2 activates viral responses in the heart repressed by INCB054329
(A) Schematic of the experiment.
(B) Lungs of SARS-CoV-2-infected K18-hACE2 mice 5 days post infection. Infection causes sloughing of bronchial epithelium, and white arrowheads show (A) collapse of alveolar spaces and (B) bronchiolar lumen.
(C) Severe lung infection with no/negligible heart infection at 4 days post infection. n = 5 mice per group.
(D) Lung RNA-sequencing reveals a robust upregulation (logFC >0.5) of 419 genes and downregulation (logFC <−0.5) of 98 genes, both FDR <0.05. n = 5 mice per group.
(E) Activation of viral responses in lungs revealed using KEGG pathway analysis of upregulated genes. Size represents number of genes regulated and the pathways of the colored circles are highlighted by the text.
(F) Stat1 and Ep300 are predicted as key transcriptional mediators of infection in the lungs.
(G) Hearts of SARS-CoV-2-infected K18-hACE2 mice 5 days post infection. Relatively normal, with no significant necrosis, fibrosis (Masson’s Tri-chrome not shown), or immune infiltrates.
(H) Heart RNA-sequencing reveals a robust upregulation (logFC >0.5) of 249 and downregulation (logFC <−0.5) of 159 genes, both FDR <0.05. n = 5 mice per group.
(I) Activation of viral responses in hearts revealed and repression of ECM using KEGG pathway analysis of upregulated genes and downregulated genes. Size represents number of genes regulated and the pathways of the colored circles are highlighted by the text.
(J) Stat1 and Ep300 are predicted as key transcriptional mediators in the heart.
(K) PCA of heart RNA-sequencing samples. n = 4–5.
(L) Heart RNA-sequencing reveals a robust upregulation (logFC >0.5) of 11 genes and downregulation (logFC <−0.5) of 91 genes, both FDR <0.05 by INCB054329. n = 4–5 mice per group.
(M) Repression of viral responses in hearts revealed using KEGG pathway analysis of downregulated genes. Size represents number of genes regulated and the pathways of the colored circles are highlighted by the text.
(N) Ep300 is predicted as the key transcriptional mediator of INCB054329 effects in the heart.
(O) Cross-analysis of the transcriptional responses in human cardiac organoids with hearts of SARS-CoV-2-infected K18-hACE2 mice.
(P) Co-regulated genes in (O) reveal a consistent activation of viral responses in both models using KEGG pathway analysis of upregulated genes. Size represents number of genes regulated, and the pathways of the colored circles are highlighted by the text.
(Q) Genes induced by both CS in human cardiac organoids and SARS-CoV-2-infected K18-hACE2 mouse hearts that are also repressed by INCB054329.
(R) Severe weight loss by 4–5 days post infection in SARS-CoV-2-infected K-18-hACE2 mice is due to severe lung infection and brain infection, and euthanasia is required.
d.p.i., days post infection; CS, cardiac cytokine storm. Data presented as mean ± SEM. ∗∗p < 0.01 using Mann-Whitney, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001 using two-way ANOVA with Sidak’s post hoc test compared to CTRL. (D, H, and L) Red dots are regulated as per the described cut-offs and gray dots are not. (F, J, and N) Values presented are adjusted p values, number of genes regulated by the transcription factor/number of genes regulated, and % of genes regulated over the total. The size of the colored slices represent the fraction of genes regulated (180° = 100%), and overlaps for each transcription factor are also depicted. Additional data bioinformatic analysis is provided in Tables S1A–S1C.