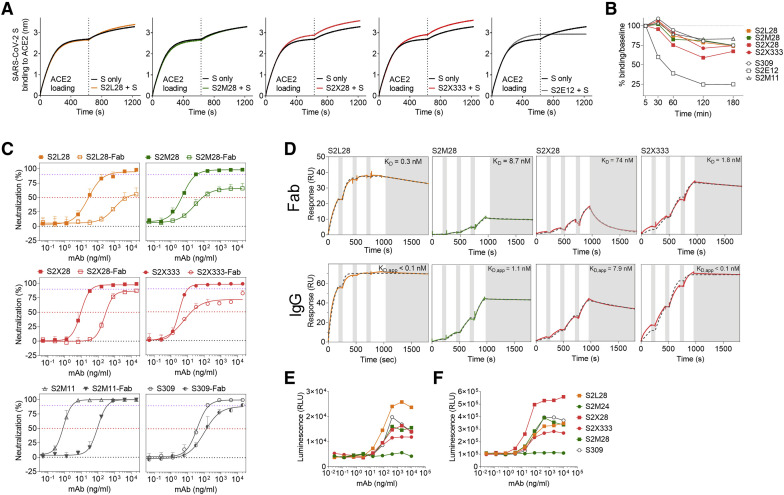
Figure 6.
Mechanism of action of NTD-specific neutralizing mAbs
(A) Competition of S2L28, S2M28, S2X28, and S2X333 with ACE2 to bind to SARS-CoV-2 S as measured by biolayer interferometry. ACE2 was immobilized at the surface of the biosensors before incubation with the S ectodomain trimer alone or pre-complexed with mAbs. The vertical dashed line indicates the start of the association of free S or S/mAb complexes with solid-phase ACE2. The anti-RBD S2E12 mAb was included as positive control.
(B) mAb-mediated S1 subunit shedding from cell surface expressed SARS-CoV-2 S as determined by flow cytometry. The anti-RBD S2M11 and S2E12 mAbs were included as negative and positive controls, respectively.
(C) Neutralization of authentic SARS-CoV-2 (SARS-CoV-2-Nluc) by S2L28, S2M28, S2X28, and S2X333 IgG or Fab. S309 and S2M11 Fab and IgG were also included as controls. Symbols are means ± SD of triplicates. Dotted lines indicate IC50 and IC90 values.
(D) SPR analysis of mAbs binding to the SARS-CoV-2 S ectodomain trimer. Gray dashed line indicates a fit to a 1:1 binding model. The equilibrium dissociation constants (KD) or apparent equilibrium dissociation constants (KD, app) are indicated. White and gray stripes indicate association and dissociation phases, respectively.
(E and F) Activation of FcγRIIa H131 (E) and FcγRIIIa V158 (F) induced by NTD-specific mAbs. The anti-RBD mAb S309 was included as positive control. One independent experiment out of at least two is shown.
See also Figures S1 and S5 and Table S1.