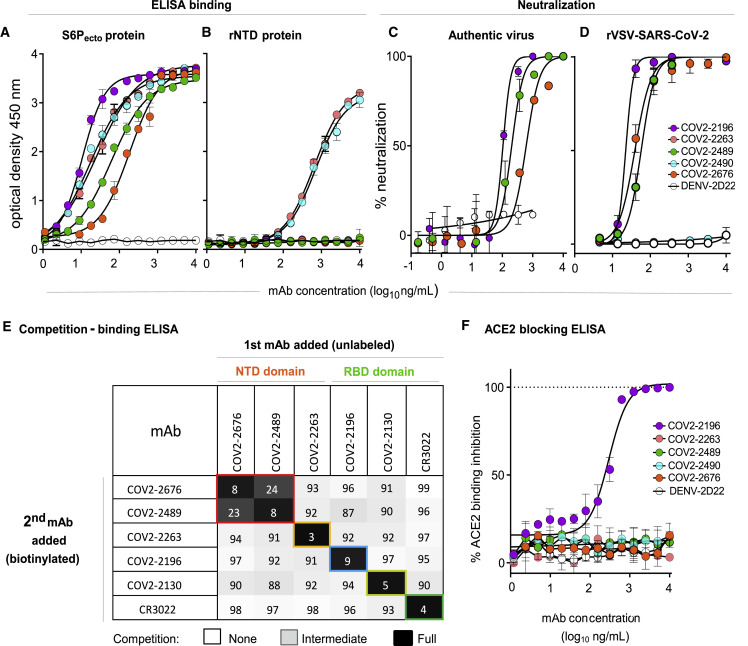
Figure 1.
Two non-RBD-reactive mAbs specific to SARS-CoV-2 S protein neutralize the virus
(A) ELISA binding of COV2-2196, COV2-2263, COV2-2489, COV2-2490, COV2-2676, or rDENV-2D22 to trimeric S-6Pecto. Data are mean ± standard deviations (SD) of technical triplicates from a representative experiment repeated twice.
(B) ELISA binding of COV2-2196, COV2-2263, COV2-2489, COV2-2490, COV2-2676, or rDENV-2D22 to rNTD protein. Data are mean ± SD of technical triplicates from a representative experiment repeated twice.
(C) Neutralization curves for COV2-2196, COV2-2489, COV2-2676, or rDENV-2D22 using wild-type SARS-CoV-2 in a FRNT assay. Error bars indicate SD; data represent at least two independent experiments performed in technical duplicate.
(D) Neutralization curves for COV2-2196, COV2-2489, COV2-2490, COV2-2676, or rDENV-2D22 in a SARS-CoV-2-rVSV neutralization assay using RTCA. Error bars indicate SD; data are representative of at least two independent experiments performed in technical duplicate.
(E) Competition binding of the panel of neutralizing mAbs with reference mAbs COV2-2130, COV2-2196, COV2-2263, COV2-2489, COV2-2676, or rCR3022. Binding of reference mAbs to trimeric S-6Pecto protein was measured in the presence of saturating concentration of competitor mAb in a competition ELISA and normalized to binding in the presence of rDENV-2D22. Black indicates full competition (<25% binding of reference antibody), gray indicates partial competition (25% to 60% binding of reference antibody), and white indicates no competition (>60% binding of reference antibody).
(F) Human-ACE2-blocking curves for COV2-2196, COV2-2263, COV2-2489, COV2-2490, COV2-2676, and rDENV-2D22 in a human-ACE2-blocking ELISA. Data are mean ± SD of technical triplicates from a representative experiment repeated twice.